Journal list menu
Export Citations
Download PDFs
ISSUE INFORMATION
ORIGINAL ARTICLE
SmartScreen-AIS: A High-Throughput qPCR Chip for Nationwide Surveillance of Aquatic Invasive Species
- First Published: 26 June 2025
Harnessing eDNA Metabarcoding to Monitor Species Diversity in Restoration Sites: Insights From Laikipia, Kenya
- First Published: 05 May 2025

The restoration of terrestrial ecosystems in Arid and Semi-arid Lands (ASALs) has gained momentum through initiatives like the Bonn Challenge and Kenya's Forest and Landscape Restoration Implementation Plan (FOLAREP). This study piloted soil environmental DNA (eDNA) metabarcoding to assess biodiversity in restored areas of Lower Naibunga Community Conservancy, Laikipia County, Kenya, using 16S rDNA and rbCL markers. Results indicated the prevalence of plant growth-promoting rhizobacteria and drought-tolerant plant species, demonstrating the potential of eDNA metabarcoding as a monitoring tool for nature-based restoration interventions.
Environmental DNA Methods for Detection of Varroa destructor in Honey Bee (Apis mellifera) Hives
- First Published: 05 May 2025

The parasitic mite, Varroa destructor, is a worldwide problem for honey bees (Apis mellifera). Using a new species-specific qPCR assay, we assessed the detection of V. destructor eDNA collected in honey and surface swabs from managed bee hives in Australia, where V. destructor has recently invaded, and in New Zealand, where V. destructor is established. We showed that these eDNA methods provide sensitive detection for V. destructor compared with a conventional bee wash method and could be a powerful complementary tool for managing the spread of this pest to new areas and detecting future incursions.
Capture—Incubate—Release: An Animal-Friendly Approach to Assess Local Aquatic Macroinvertebrate Species Diversity Through Environmental DNA Metabarcoding
- First Published: 05 May 2025
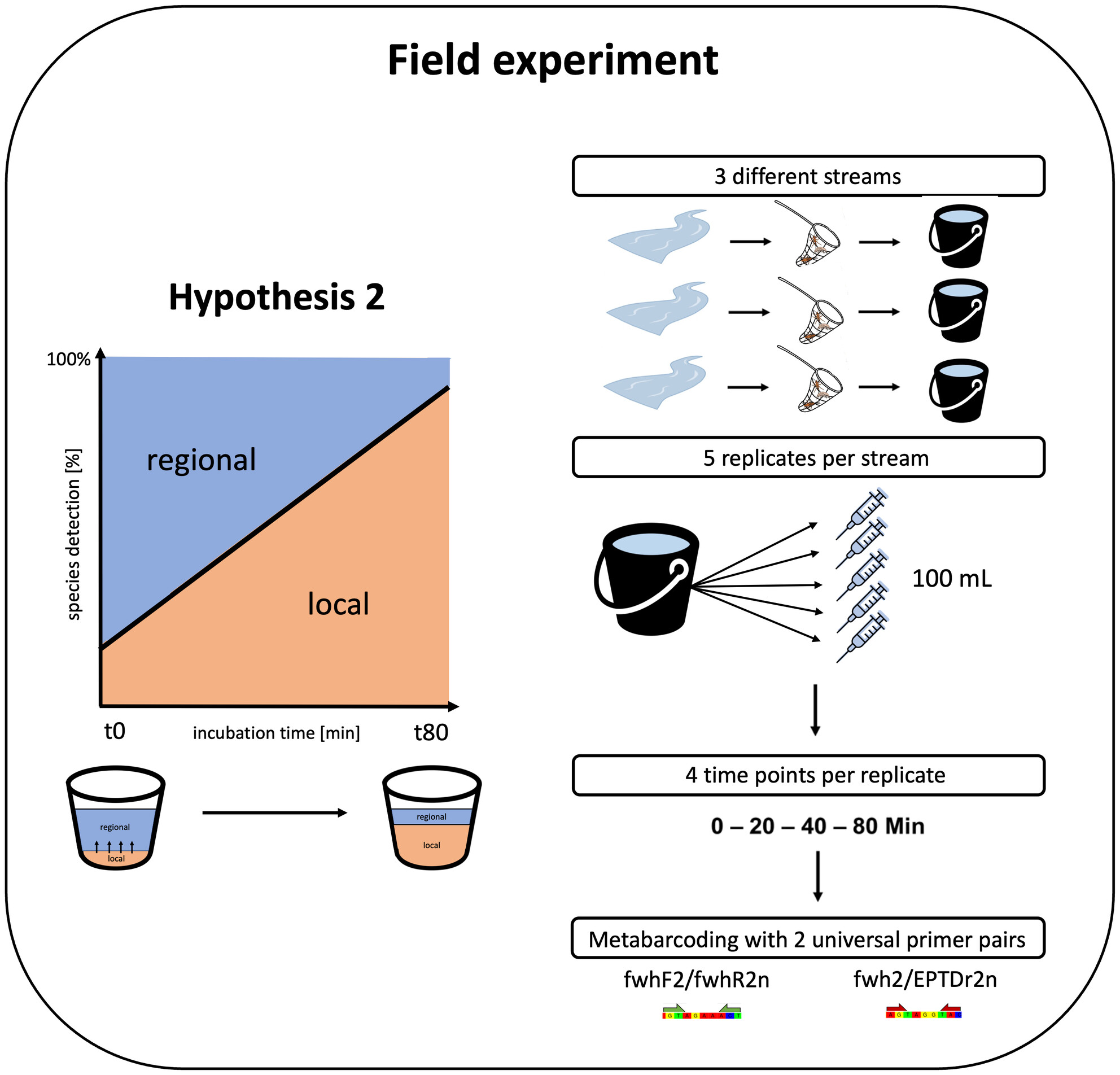
We assessed whether we can combine the advantages of both bulk and eDNA metabarcoding, that is, obtaining a local signal and being minimally invasive, to improve local invertebrate detections in streams using water samples. We quantified eDNA release over time after a short incubation in water for single species using qPCR and for bulk samples using metabarcoding. Our results showed a much higher overlap between bulk and enriched eDNA metabarcoding (55%–60%) in comparison to reported overlaps between bulk and stream eDNA metabarcoding from other studies (often < 20%). However, this overlap did not increase with incubation time.
METHOD
Automating the Curation of DNA Barcode Databases for Vascular Plants
- First Published: 26 June 2025
ORIGINAL ARTICLE
In Vitro Passive eDNA Sampling Provides a Cost-Effective Alternative for Large Scale Sample Collection
- First Published: 20 May 2025
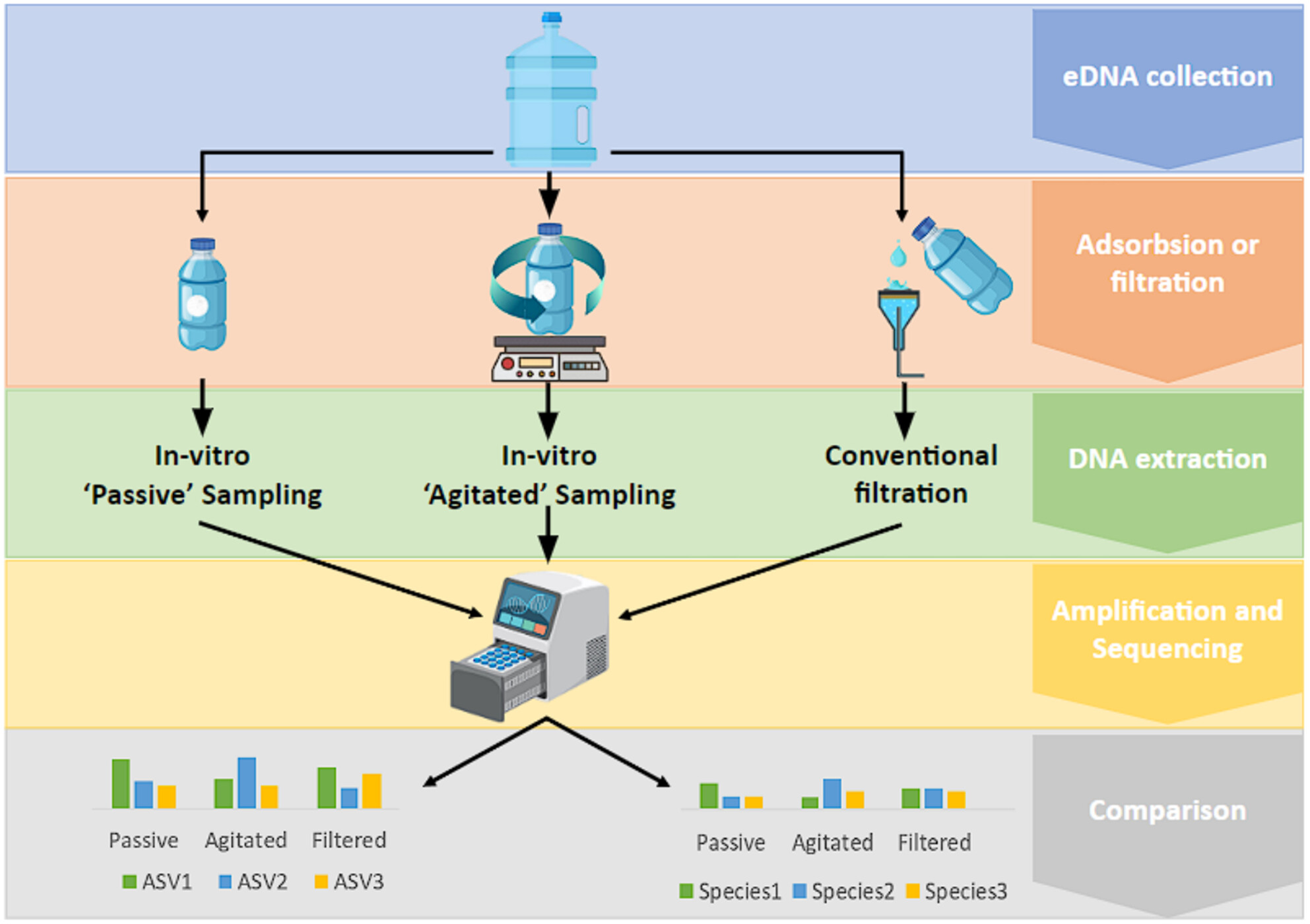
Here we trial new passive eDNA sampling methods, which we term ‘in vitro passive eDNA sampling’; where a water aliquot is collected at a sampling location and passively sampled in a microcosm experiment, allowing for high-density sampling while the researcher moves to other areas. We show that at a species level, in vitro passive sampling methods can return comparable species richness estimates to conventional filtration at both estuarine and inshore marine locations. In addition, agitating our in vitro passive samples improved DNA yields by an average of 2.2× as opposed to in vitro passive sampling without agitation, though both were significantly lower than filtration.
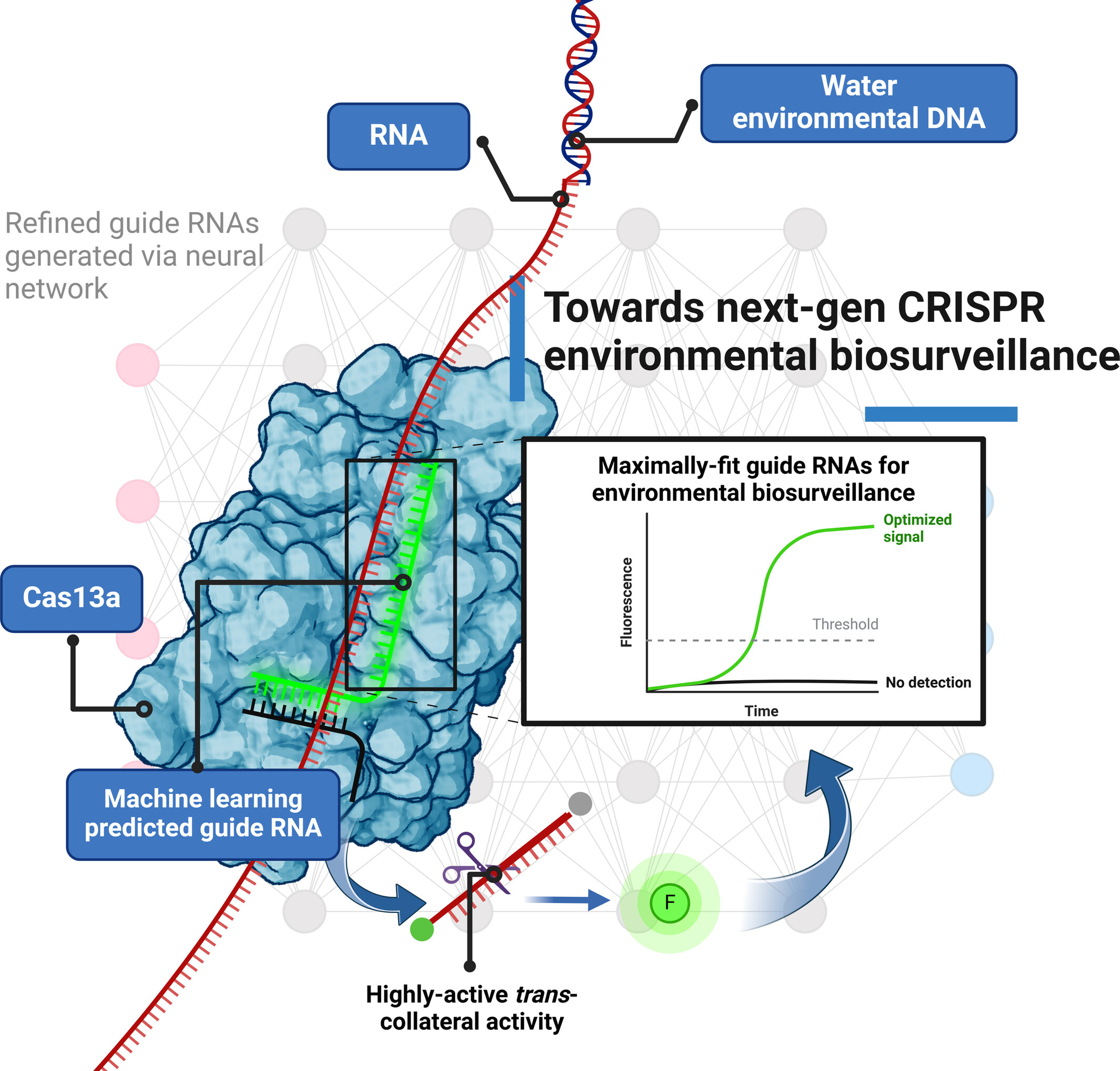
CRISPR-Based Environmental Biosurveillance Assisted via Artificial Intelligence Design of Guide-RNAs
- First Published: 09 May 2025
Drivers of Insect Diversity and Community Turnover in Protected Tropical Deciduous Forests of Mexico
- First Published: 18 June 2025
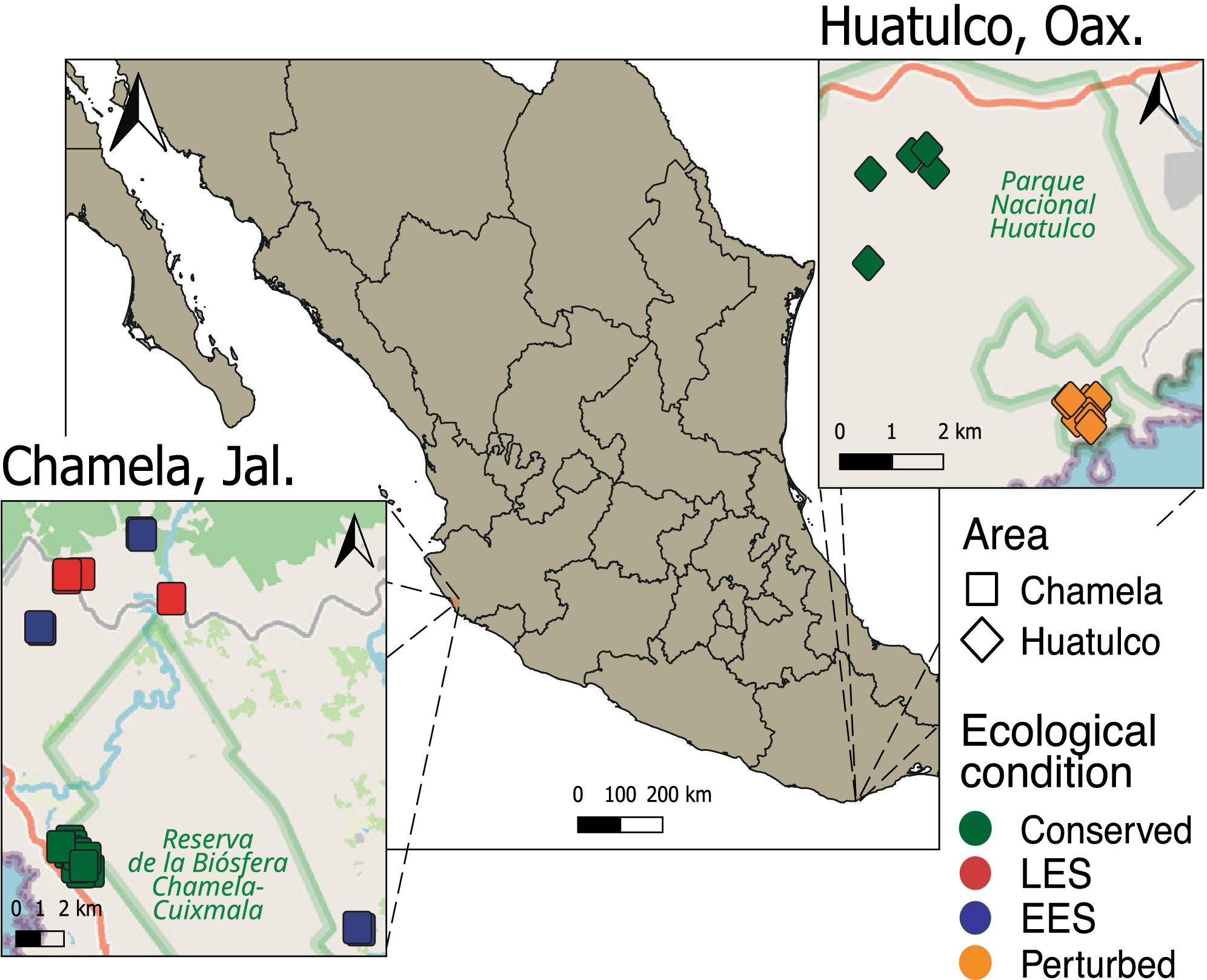
Seasonal fluctuations and anthropogenic disturbance drive insect diversity and community structure in two protected tropical deciduous forests on Mexico's Pacific coast. High regional turnover in insect communities underscores the unique biodiversity of tropical deciduous forests. Habitat features, including deadwood, tree diversity, and canopy coverage, also shape local insect diversity and community turnover.
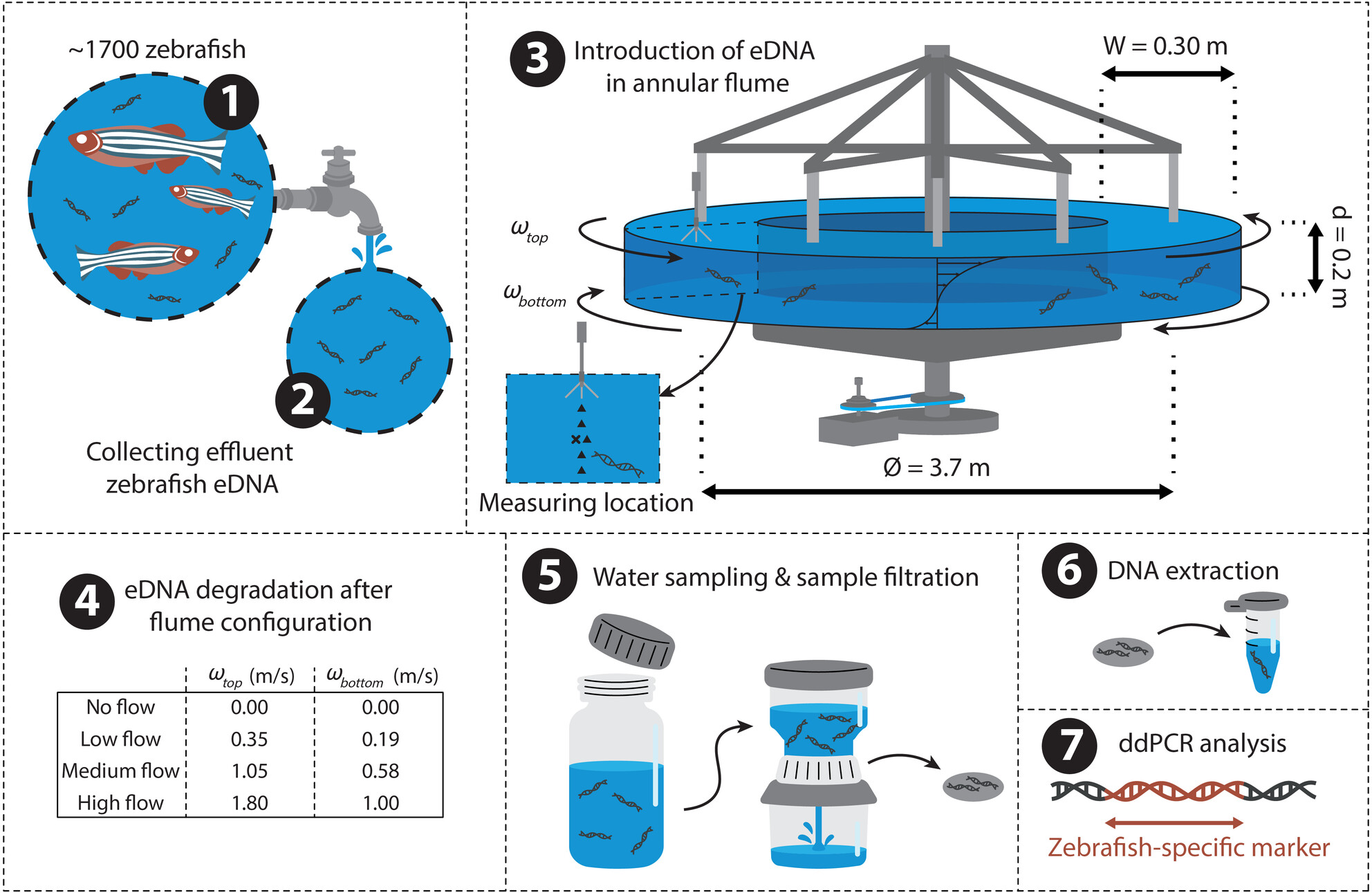
The Impact of Flow Velocity on Environmental DNA Detectability for the Application in River Systems
- First Published: 07 May 2025
Tropical Rainfall and eDNA Washout Impact Estimations of Amazonian Biodiversity Patterns From Environmental Samples
- First Published: 17 June 2025
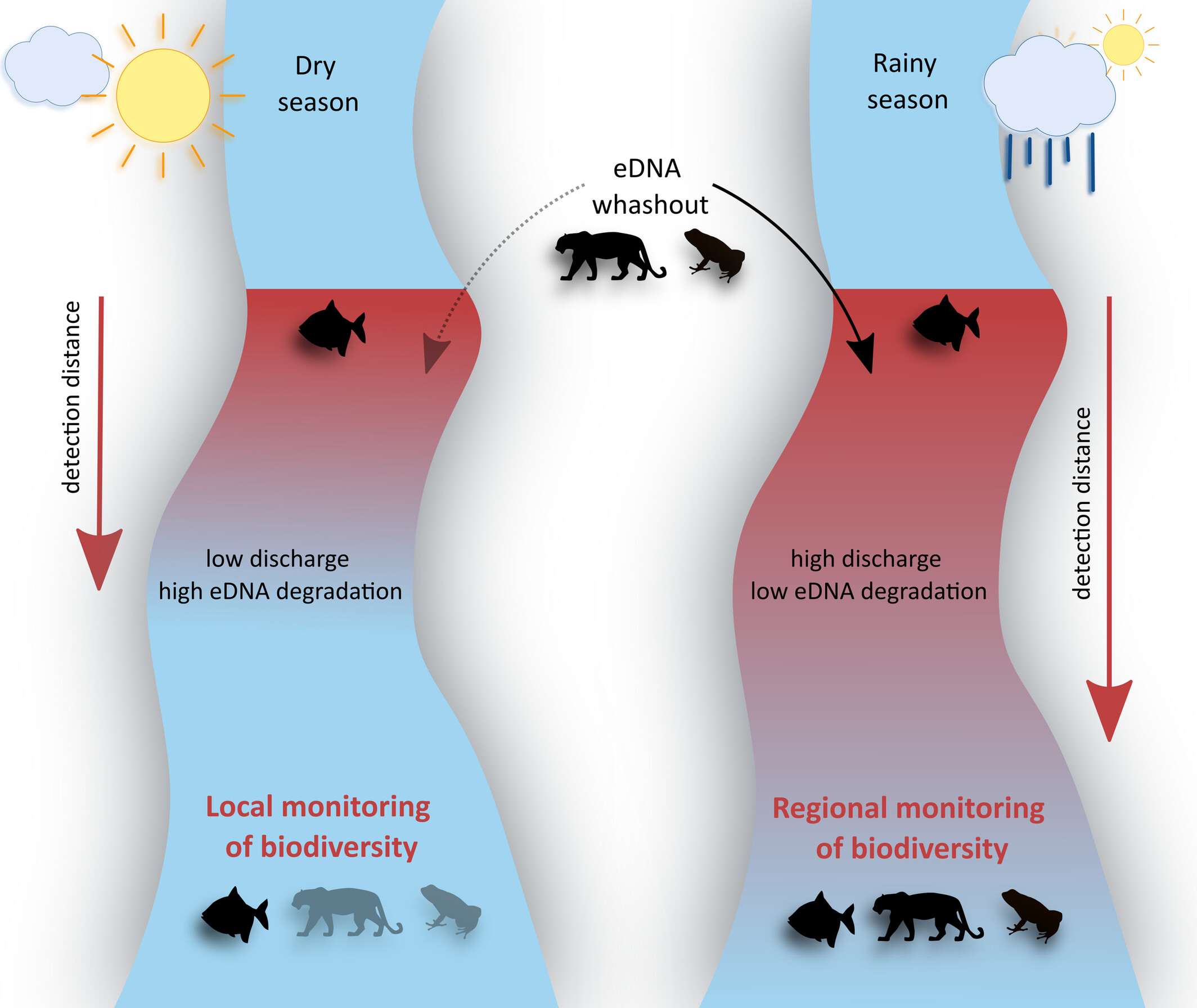
We used eDNA to assess seasonal biodiversity patterns in a tropical river in French Guiana. Our results show that rainfall increases the detection of terrestrial species through eDNA washout and leads to species homogenization via increased eDNA transport. These findings highlight how seasonal changes can shape eDNA signals, with implications for optimizing sampling strategies.
DNA Extraction Methodology has a Limited Impact on Multitaxa Riverine Benthic Metabarcoding Community Profiles
- First Published: 09 May 2025
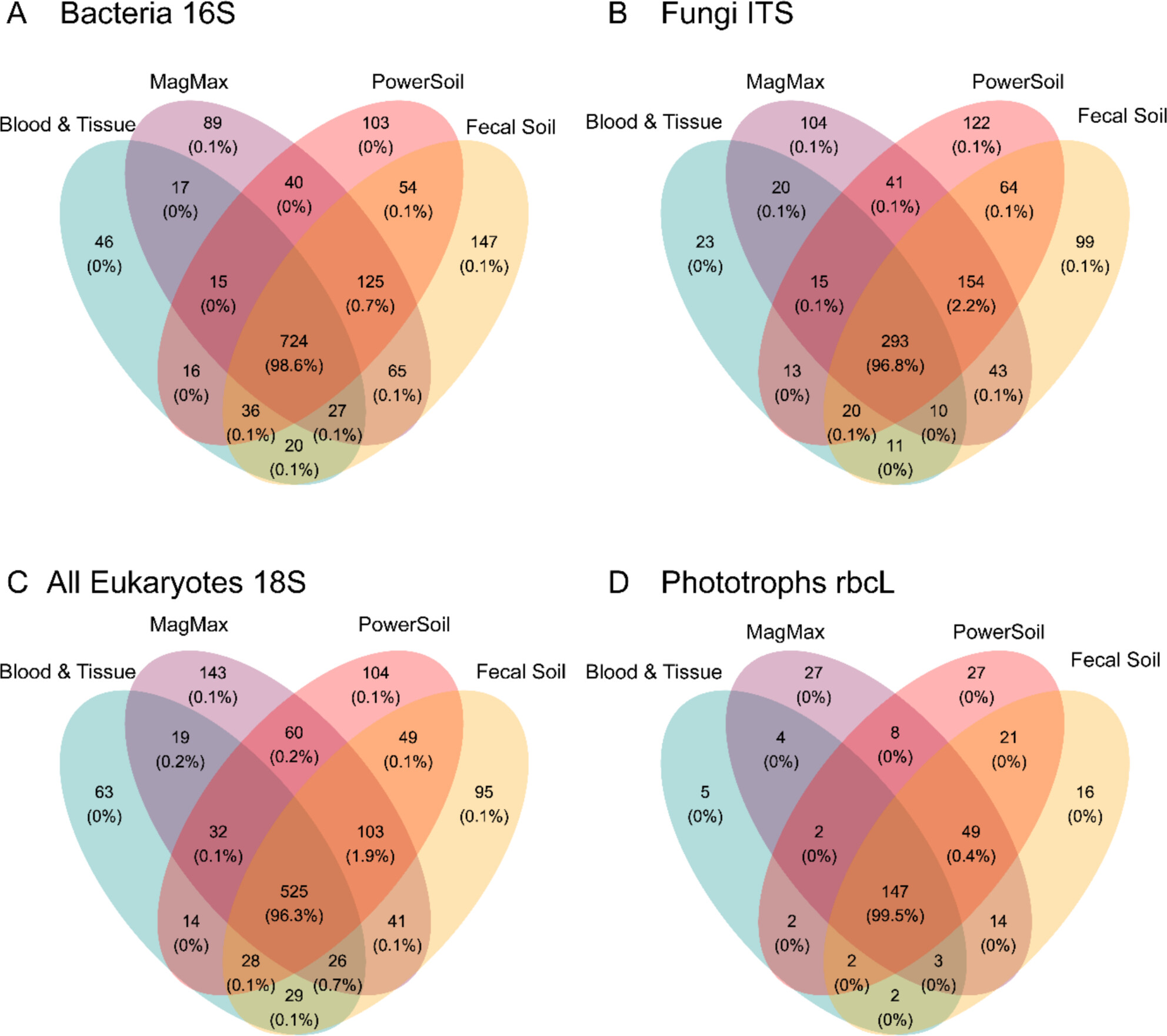
This work examines how DNA extraction methodology may bias the analysis of community composition using amplicon sequencing at a cross-kingdom level in river phytobenthos samples. Our overall findings suggest that, irrespective of kit, the application of mechanical lysis is likely to improve low abundance community member detection of microbes. However, the high correlation between sample origin, irrespective of the extraction method employed, would imply that existing data collected using alternative methodologies are still valid to include and inform future monitoring practices.
Distinct Community Compositions of Prokaryotes, Eukaryotes, and Fishes Revealed Through Environmental DNA Analysis at Different Salinities Around Ishigaki Island, Okinawa, Japan
- First Published: 17 June 2025
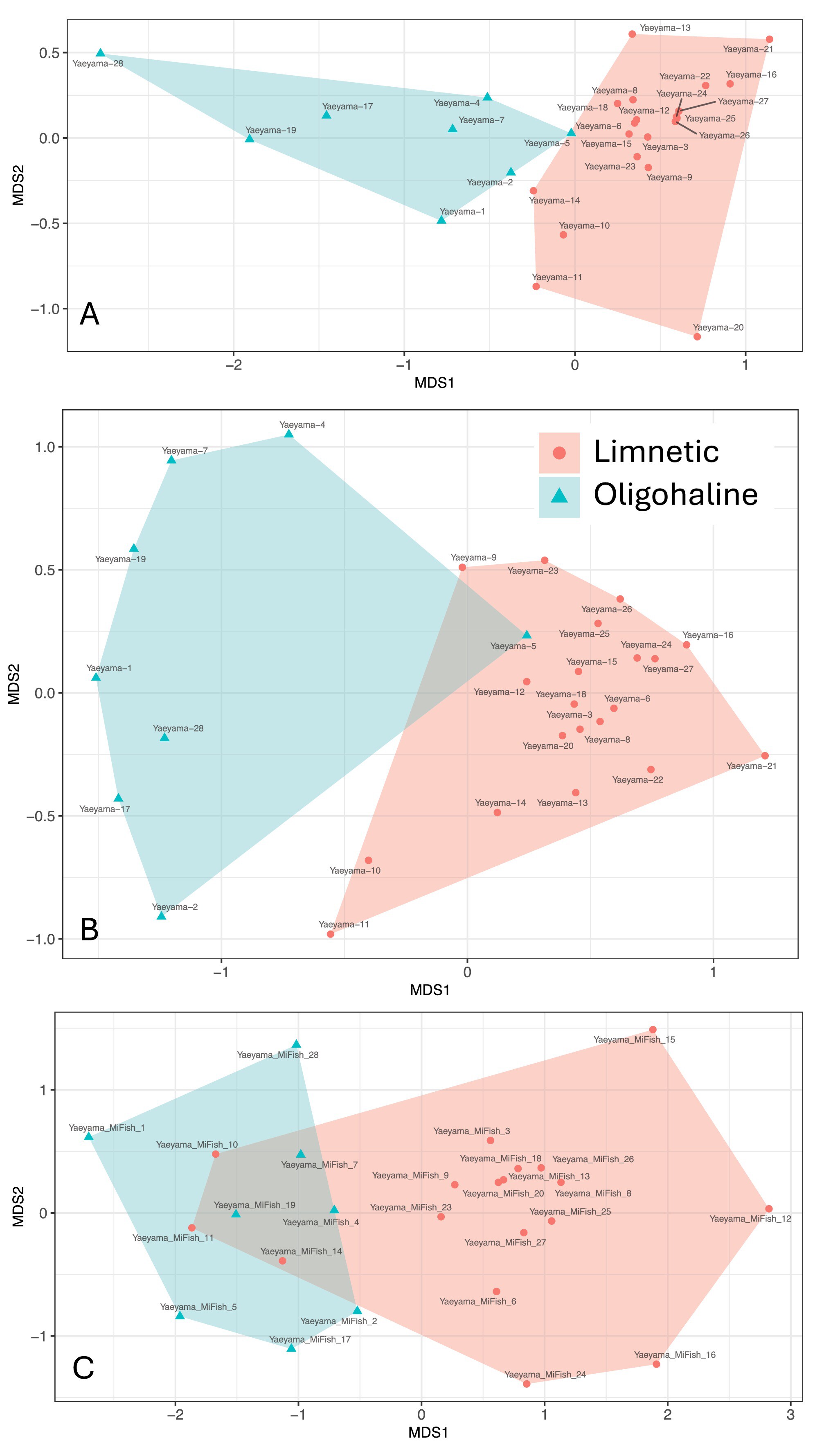
Multi-level eDNA metabarcoding revealed that even slight salinity changes alter the community composition of microorganisms and fish. The effect of salinity on alpha-diversity was taxa-dependent. Some potentially harmful microorganisms for fish were found, highlighting the importance of utilizing eDNA for monitoring.
Detection of Non-Native Pink Salmon (Oncorhynchus gorbuscha) in Swedish Rivers Using eDNA
- First Published: 12 May 2025
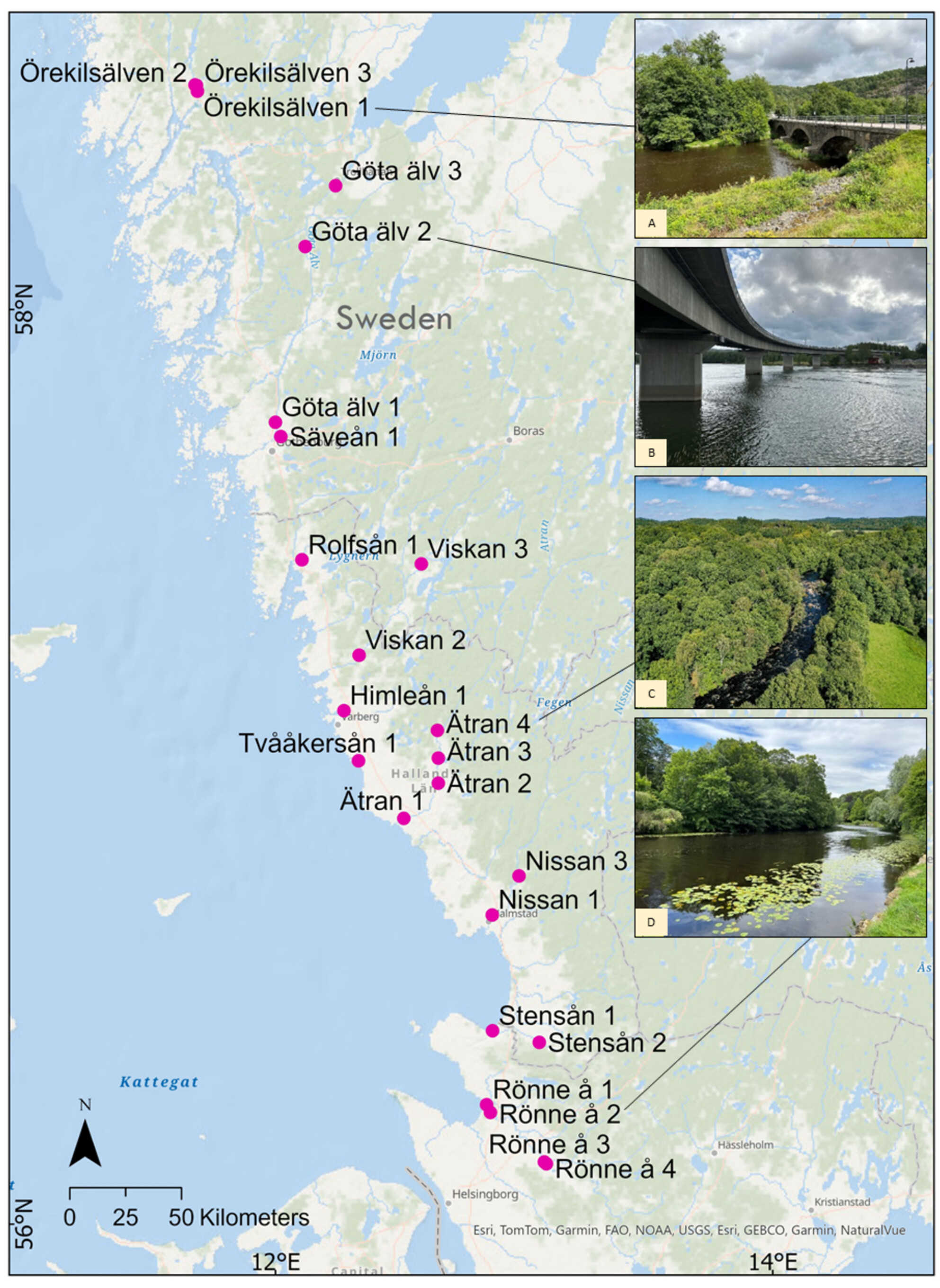
eDNA surveys were conducted in 27 river systems in south-western Sweden in 2023 to detect adult spawning pink salmon. Results indicated, through mainly qPCR methods, the presence of pink salmon at 24 sites across 11 river systems along the West Coast. Pink salmon were not detected in any of the sampled rivers that drain into the Baltic Sea; however, there is a high risk that they will spread into this region in the coming years.
eDNA Replicates, Polymerase and Amplicon Size Impact Inference of Richness Across Habitats
- First Published: 09 May 2025
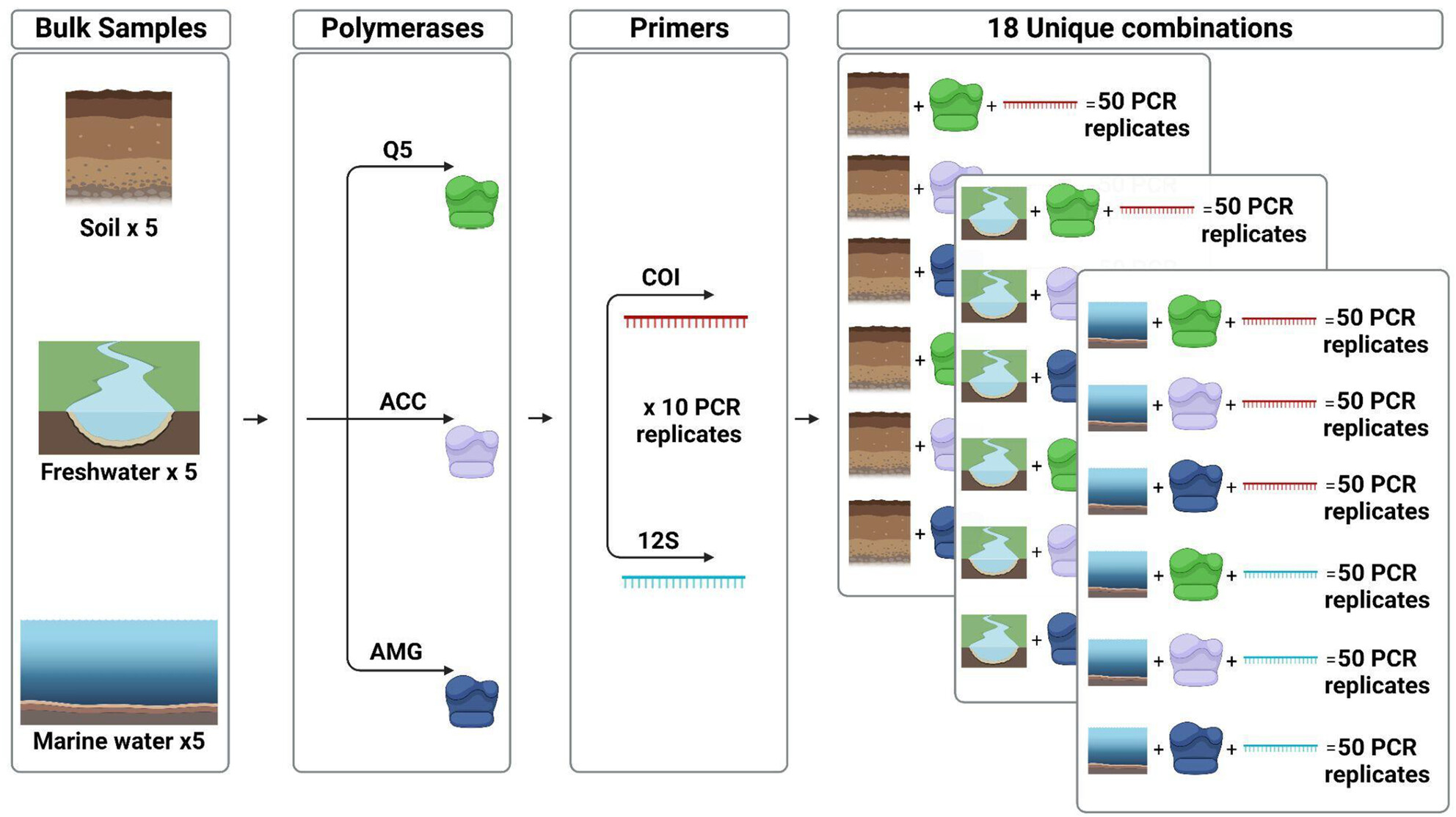
We conducted a large and controlled experiment, with bulk samples from terrestrial, freshwater and marine environments, to investigate the impacts of natural and technical replicate numbers, PCR polymerase and amplicons sizes on the recovered richness. We found variations in the recovered richness depending on PCR polymerase choice and highlight the dissimilar impacts between natural and technical replicates. We highlight the benefits and limitations of replication strategies, polymerase choice and amplicon size across terrestrial, marine and freshwater habitats, and provide recommendations to increase the reliability of future eDNA-based metabarcoding studies.
METHOD
Environmental DNA (eDNA) Quantitative Polymerase Chain Reaction-Based Assays for Surveying 125 Taxa of Importance to North America
- First Published: 12 June 2025
We present 125 eDNA quantitative real-time polymerase chain reaction (qPCR) assays against animal species of importance to North America. We show how these assays were designed and validated to comply with the Canadian national eDNA standards to allow confident eDNA detection for coastal and inland ecological surveys and biosurveillance.
ORIGINAL ARTICLE
Decaying Uncertainties: Exploring the Role of Decay Rate Variability in Marine eDNA Dispersal Using Lagrangian Transport Modeling
- First Published: 10 June 2025
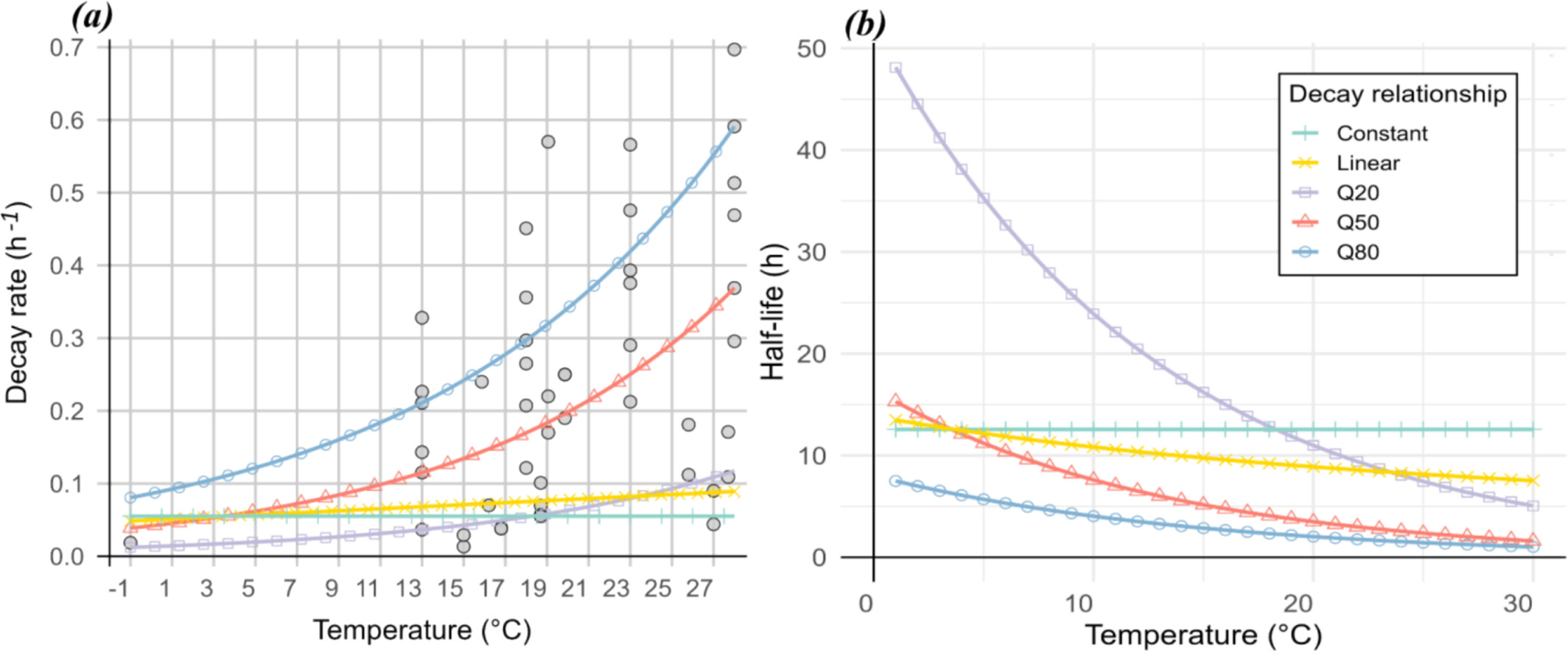
Environmental DNA is a powerful tool for biodiversity monitoring. Using a Lagrangian particle-tracking model, we examined how different temperature-dependent decay relationships affect eDNA transport and persistence in the Bay of Biscay. Our results show that current velocity was the primary driver of transport, while decay rate had a stronger influence on dispersion and lifetime. In contrast, temperature had minimal impact on transport variability.
Out in the Open: Investigating Passive Airborne eDNA Detection of Bats at Artificial Feeding Stations
- First Published: 09 May 2025
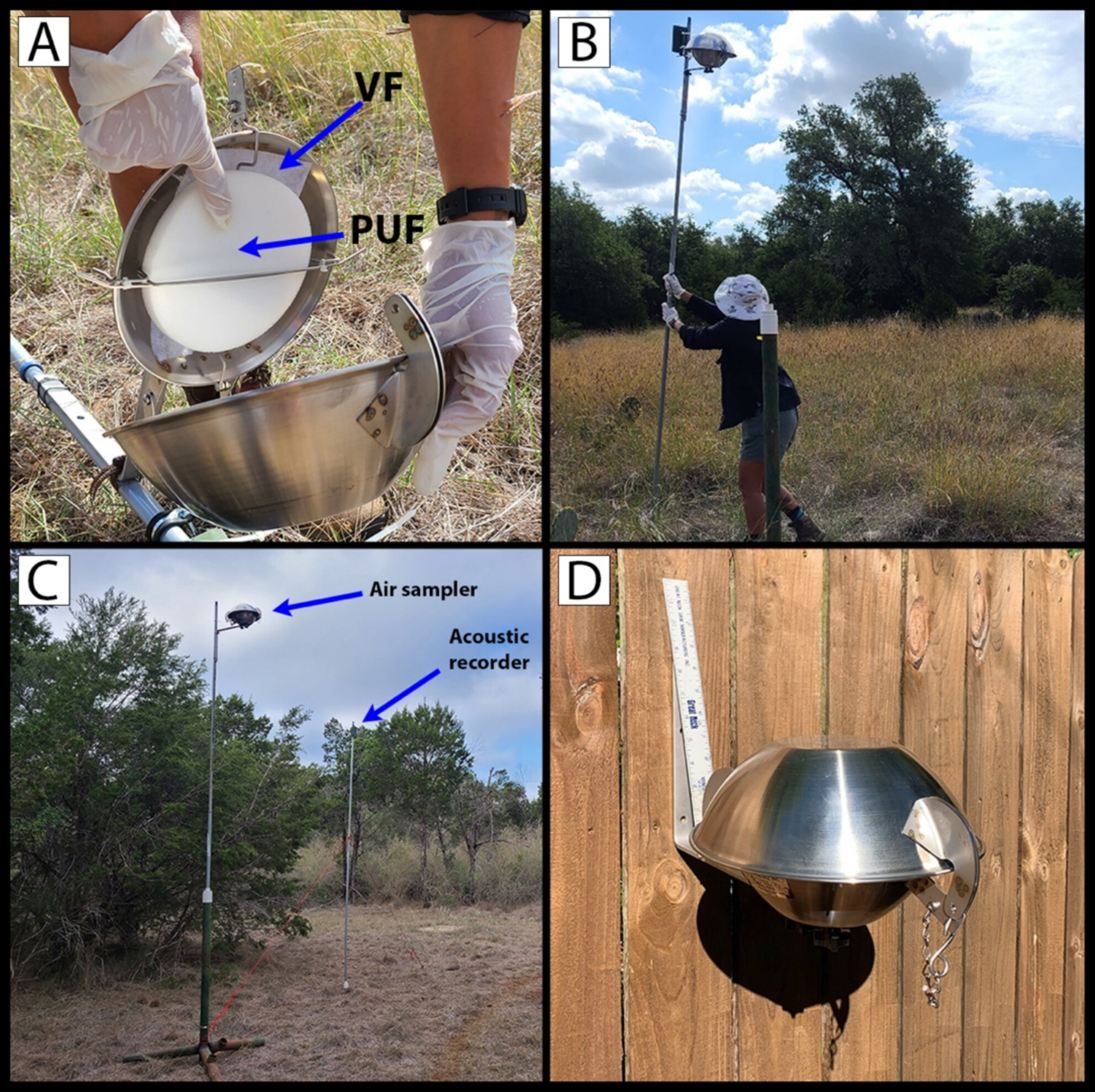
This study employs passive air sampling to detect environmental DNA (eDNA) of bats using DNA metabarcoding and a novel qPCR assay for the imperiled tricolored bat (Perimyotis subflavus). Metabarcoding successfully allowed detection of two bat species in one sample, and qPCR allowed detection of the tricolored bat in two samples, providing a lower bound possibility for open-air detection of bats using passive air sampling but highlighting the need for improved sampling methods.
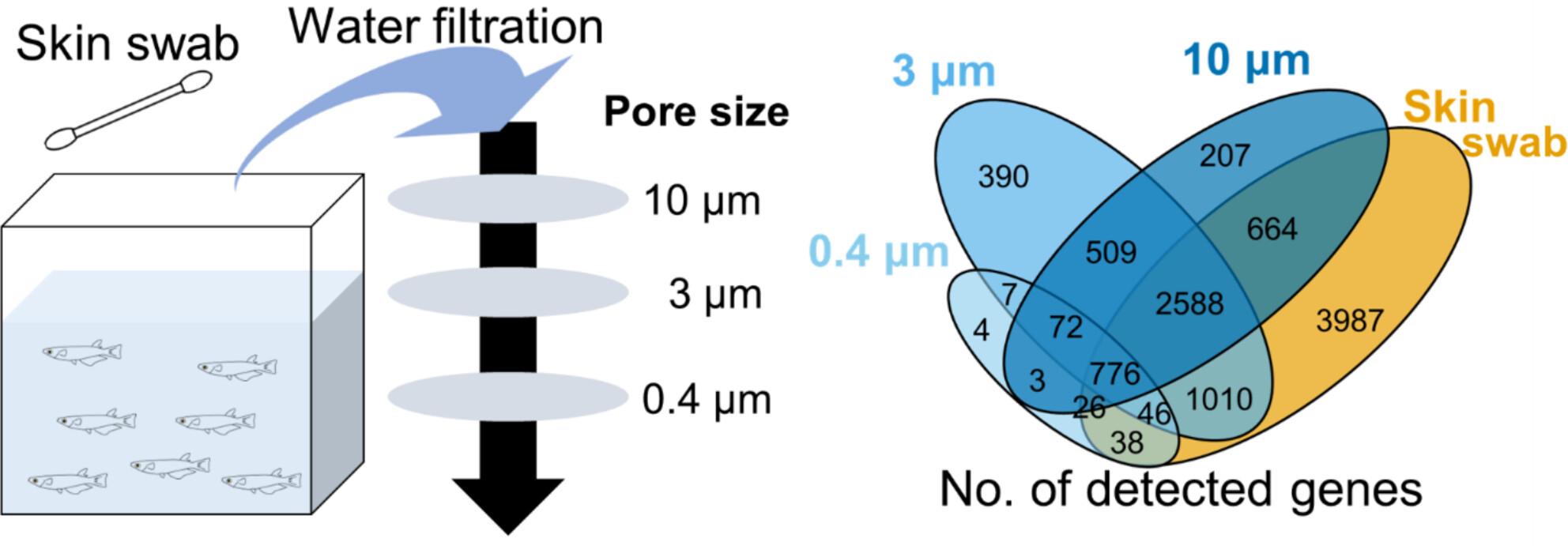
Comprehensive Sequencing of Environmental RNA From Japanese Medaka at Various Size Fractions and Comparison With Skin Swab RNA
- First Published: 10 June 2025
Water Interaction Type Affects Environmental DNA Shedding Rates of Terrestrial Mammal eDNA Into Surface Water Bodies
- First Published: 10 June 2025
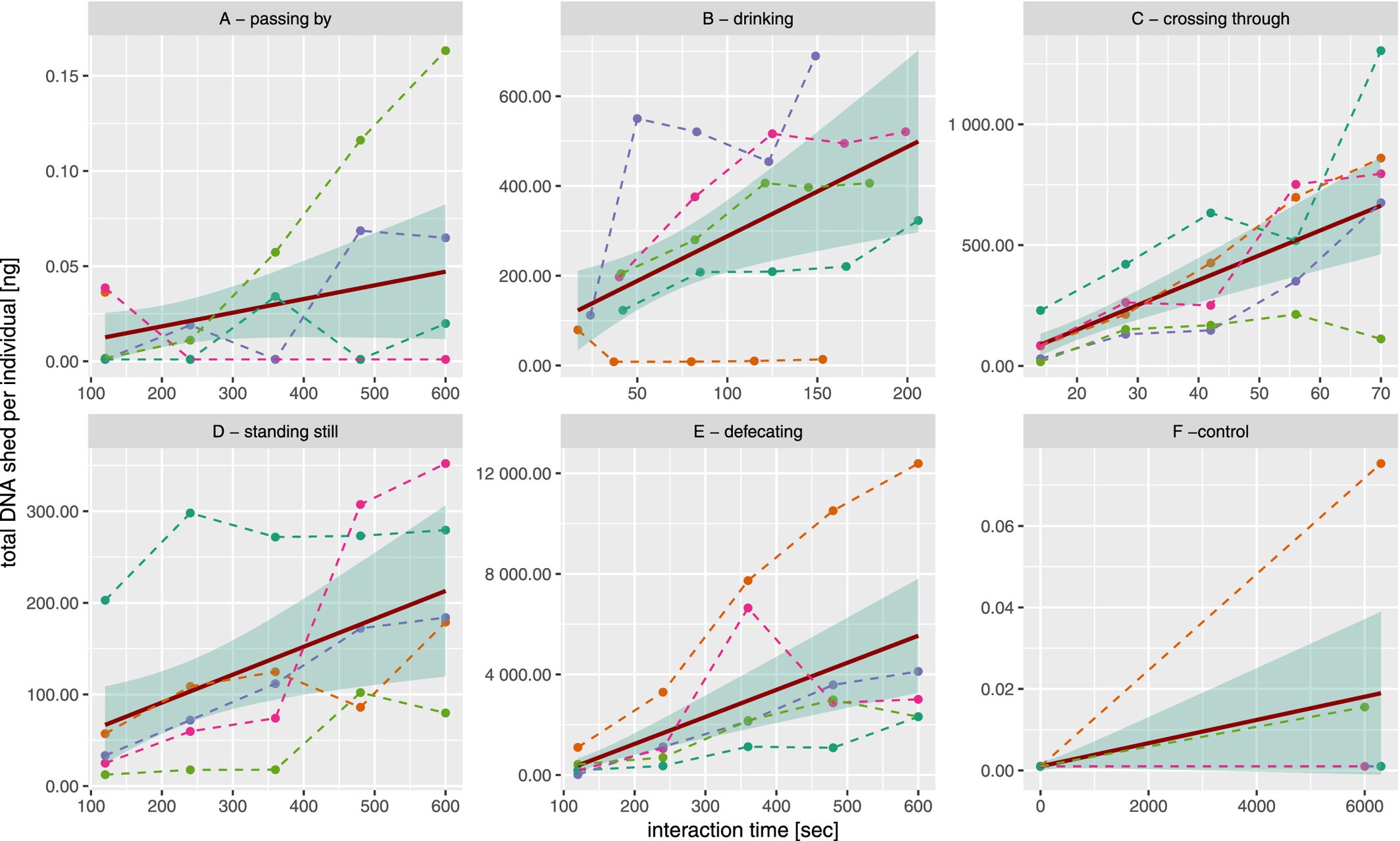
Although shedding rates for aquatic species are well studied, little is known about terrestrial mammal shedding rates in water. In this study, we quantify eDNA shedding rates from domestic dogs during various interactions with water bodies and find that defecation and crossing through water produce the highest shedding rates, highlighting direct water interaction as an important factor for eDNA deposition and detection probabilities.
Geographic Genetic Divergence in Tychoplanktonic Taxa Dominating Diatom Communities in Marine Biofilms
- First Published: 10 June 2025
Improving the Understanding of Detections From iDNA Surveys in Malaysian Borneo With Multiscale Occupancy Models: A Case-Study Using Leech Blood Meals
- First Published: 19 May 2025
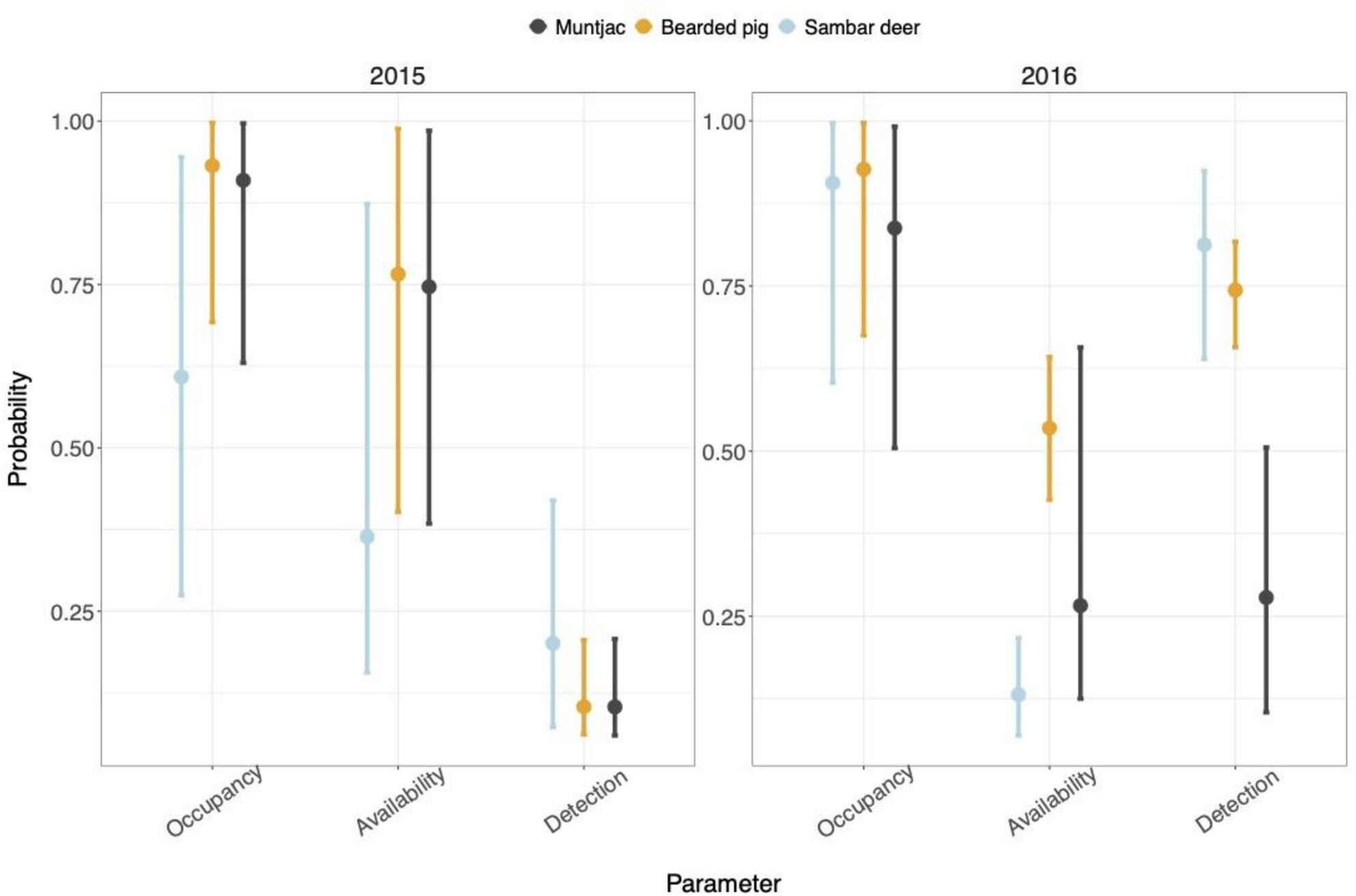
Here we reanalyse an invertebrate-derived DNA (iDNA) survey of mammal detections from across a habitat gradient in Sabah, Malaysian Borneo, using multiscale occupancy models. First we estimate three probabilities describing the occupancy, availability, and detection of three abundant mammals (bearded pig, muntjac and sambar deer) and then investigate how these are impacted by environmental, sampling and technical covariates. Our results highlight the species and season specific responses to these variables and the trade-off between maximising probabilities detection and realistic amounts of sampling.
Environmental DNA Documents Ecosystem-Wide Biodiversity Within the Marine Protected Area Stellwagen Bank National Marine Sanctuary
- First Published: 09 June 2025
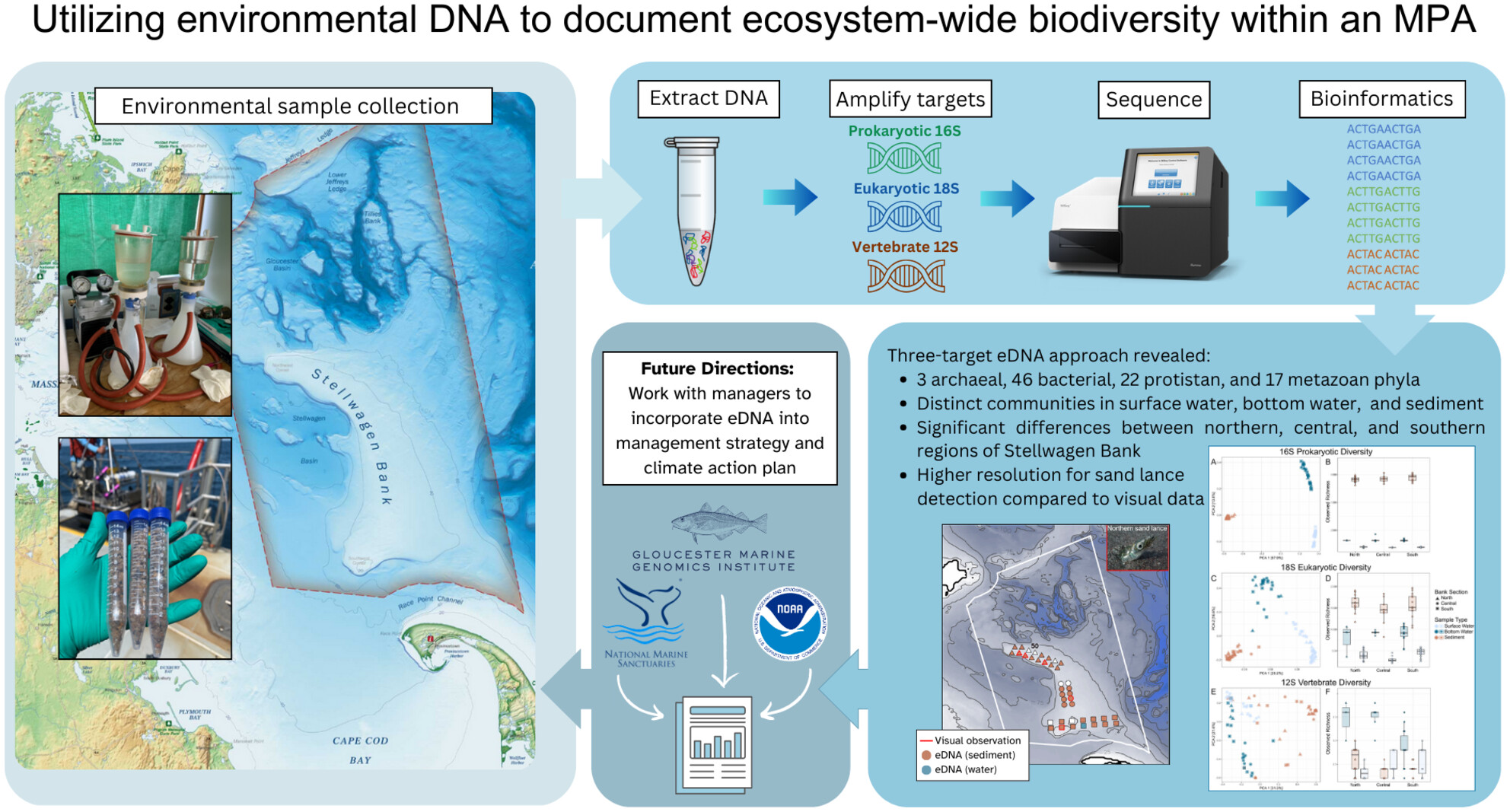
Three marker genes targeting prokaryotic, eukaryotic, and vertebrate diversity were used to assess biodiversity within Stellwagen Bank National Marine Sanctuary. High levels of biodiversity were seen across the sampled area, with distinct communities across surface water, bottom water, and sediment. When compared to traditional visual data, eDNA provided higher resolution habitat use data for the Northern sand lance.
Quantifying the Sensitivity of Targeted eDNA Surveys to Improve Detection of Invasive Cane Toads (Rhinella marina)
- First Published: 07 June 2025
Survey sensitivity is an important consideration for designing monitoring programs, though quantifying the sensitivity of eDNA surveys and examining the factors influencing this in the field remain understudied. Here we quantify the effect of key factors on eDNA sensitivity and compare the performance of eDNA surveys to conventional visual surveys for detecting invasive cane toads (Rhinella marina) across their invasion front. By doing so, we show how eDNA surveys can be tailored to achieve a specified level of detection probability at individual sites given variation in factors that affect sensitivity (e.g., toad density, size of the water body sampled).
eDNA-Based Detection of Invasive Crayfish and Crayfish Plague in Estonia
- First Published: 20 May 2025
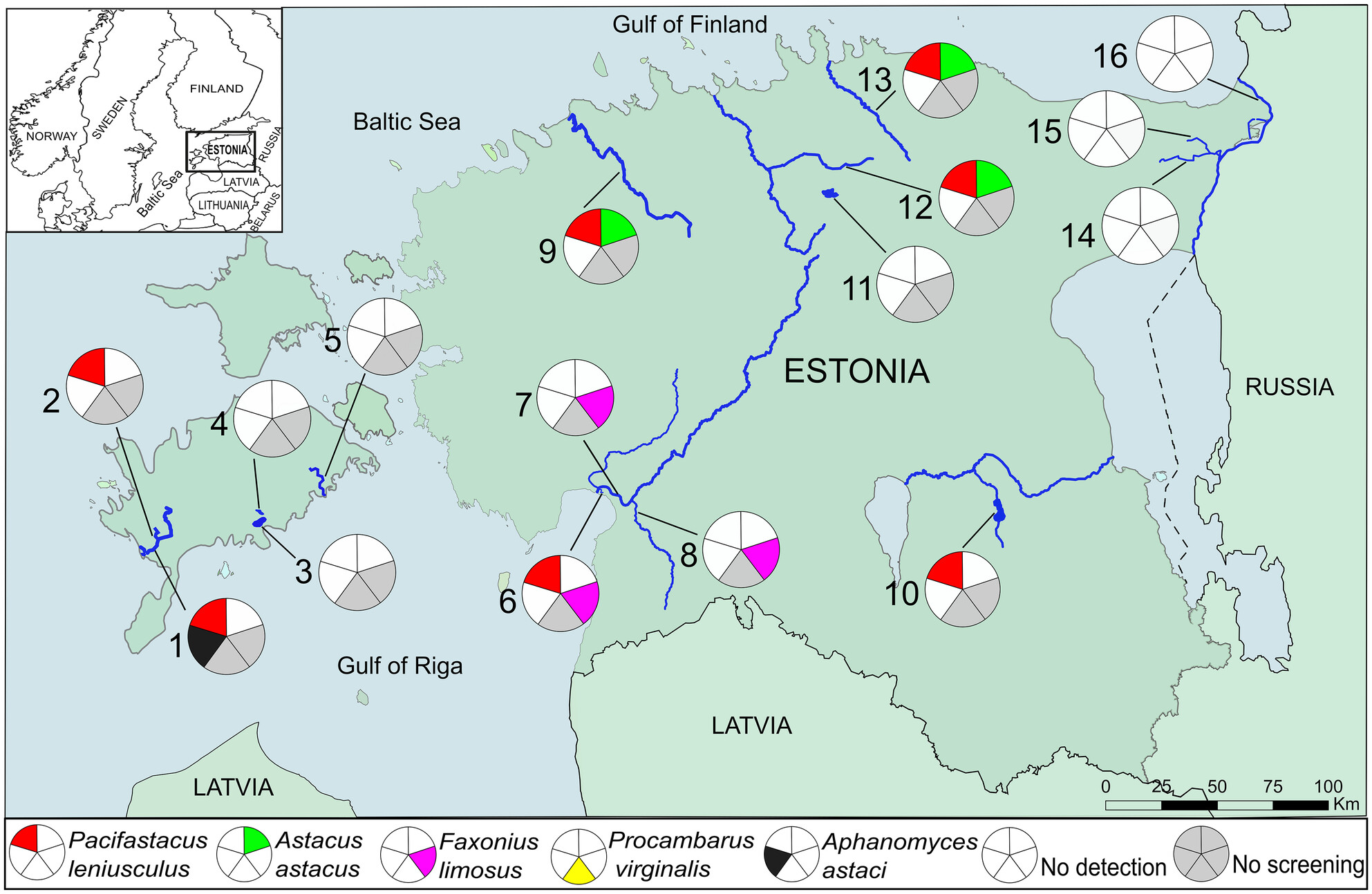
Our study explored the potential of integrating the eDNA approach into ongoing annual monitoring programs for invasive crayfish species and A. astaci. Crayfish eDNA was detected in 9 out of 14 water bodies where presence was confirmed by trapping, yielding 64% detection efficiency. eDNA from A. astaci was found in only one water body hosting invasive crayfish species. With further development, the eDNA approach can be integrated into routine monitoring of crayfish and crayfish plague pathogen as a supplement to trapping.
Promoting Community-Led Monitoring of Taonga (Treasured) Species and Freshwater Health Through eDNA Metabarcoding
- First Published: 20 May 2025
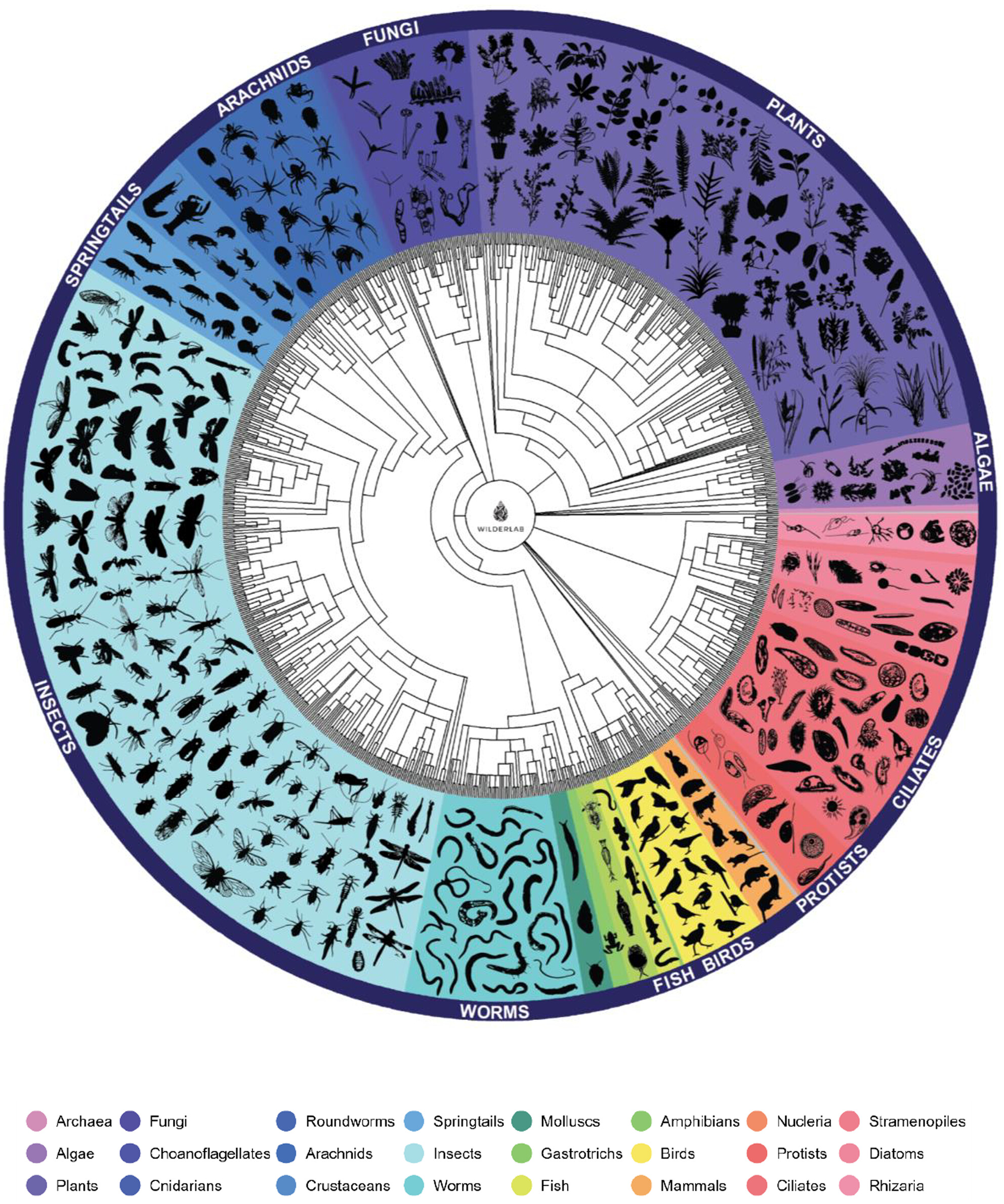
The manuscript describes our partnership with a rural community and the use of freshwater tree-of-life eDNA analysis methods to provide a tangible demonstration of the ecosystem health and biodiversity measurements of their local water catchment. The eDNA methods were able to provide a clear link back to culturally significant species, some of which were readily familiar not only to the local community but also to other species that had not been observed prior. In addition, this study took a novel approach to investigate the link between conventional water quality assessment parameters (e.g., Escherichia coli, total nitrogen, turbidity, conductivity, etc.) across contrasting land uses and demonstrate association with the taxon-independent community index (TICI), a new ecosystem health metric, and to eDNA reads from fecal source species. This study will assist water managers with improved understanding of the utility of eDNA methods for ecosystem health and biodiversity assessments and provide a framework from which catchment communities are able to reconnect with their local landscapes.
PERSPECTIVE
A Metadata Checklist and Data Formatting Guidelines to Make eDNA FAIR (Findable, Accessible, Interoperable, and Reusable)
- First Published: 06 June 2025
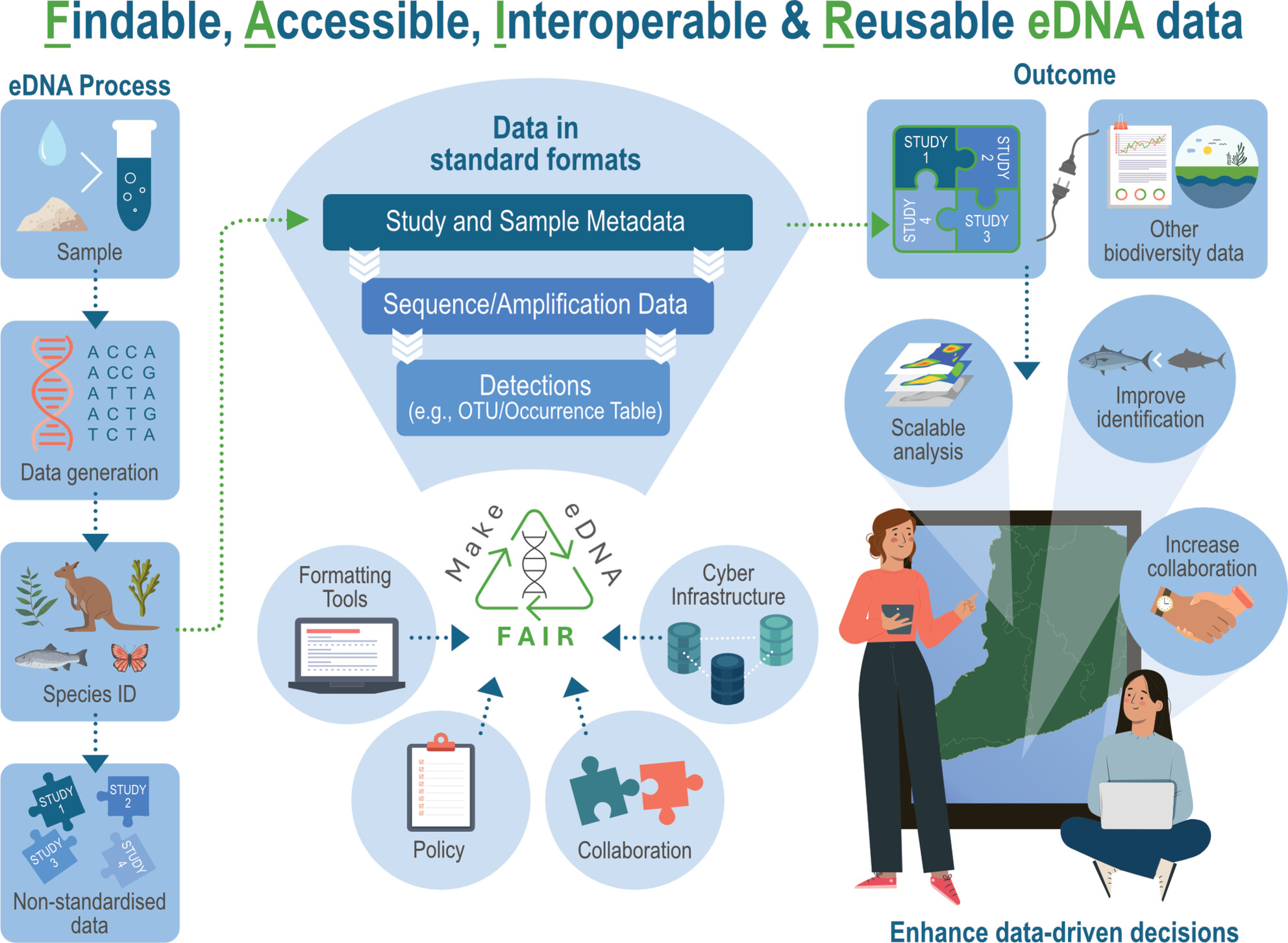
Environmental DNA (eDNA) has emerged as a transformative tool for biodiversity monitoring and species detection, yet inconsistent metadata practices hinder data interoperability and reuse. To address these challenges, we present the comprehensive FAIR eDNA (FAIRe) guidelines and metadata checklist, supported by practical tools and multi-disciplinary collaborations, to advance the adoption of FAIR (Findable, Accessible, Interoperable, and Reusable) principles in eDNA workflows.
ORIGINAL ARTICLE
Environmental Gradients, Not Geographic Boundaries, Structure Meiofaunal Communities in Siberian Seas
- First Published: 23 May 2025
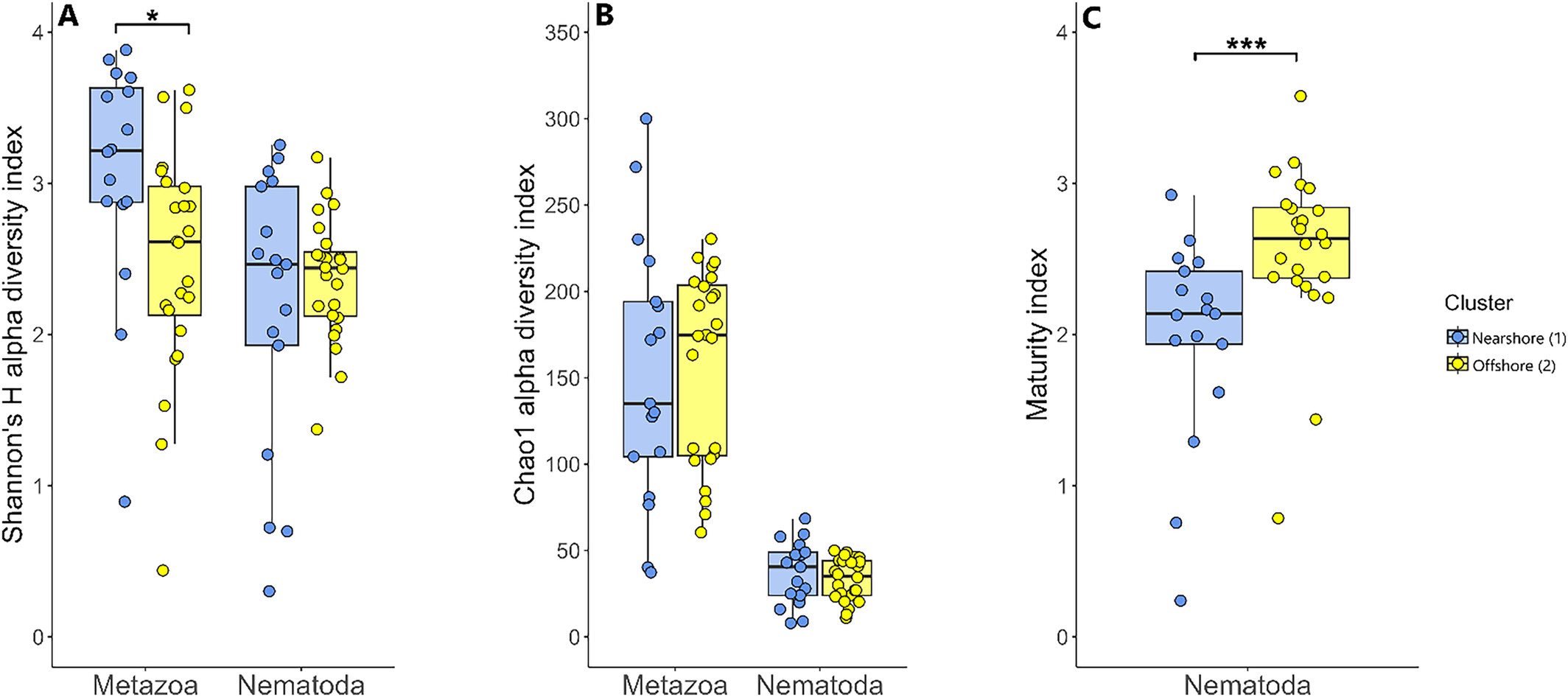
This study uses sediment eDNA metabarcoding to assess the diversity and distribution of meiofauna in Siberian seas, marking the first application of this technique in the region. Our results reveal that meiofauna diversity is similar across the three studied seas; however, both meiofauna diversity and the maturity index of nematode communities are significantly affected by coastal erosion and river discharge at nearshore stations. As climate change progresses, these factors are anticipated to intensify, further influencing meiofaunal communities.
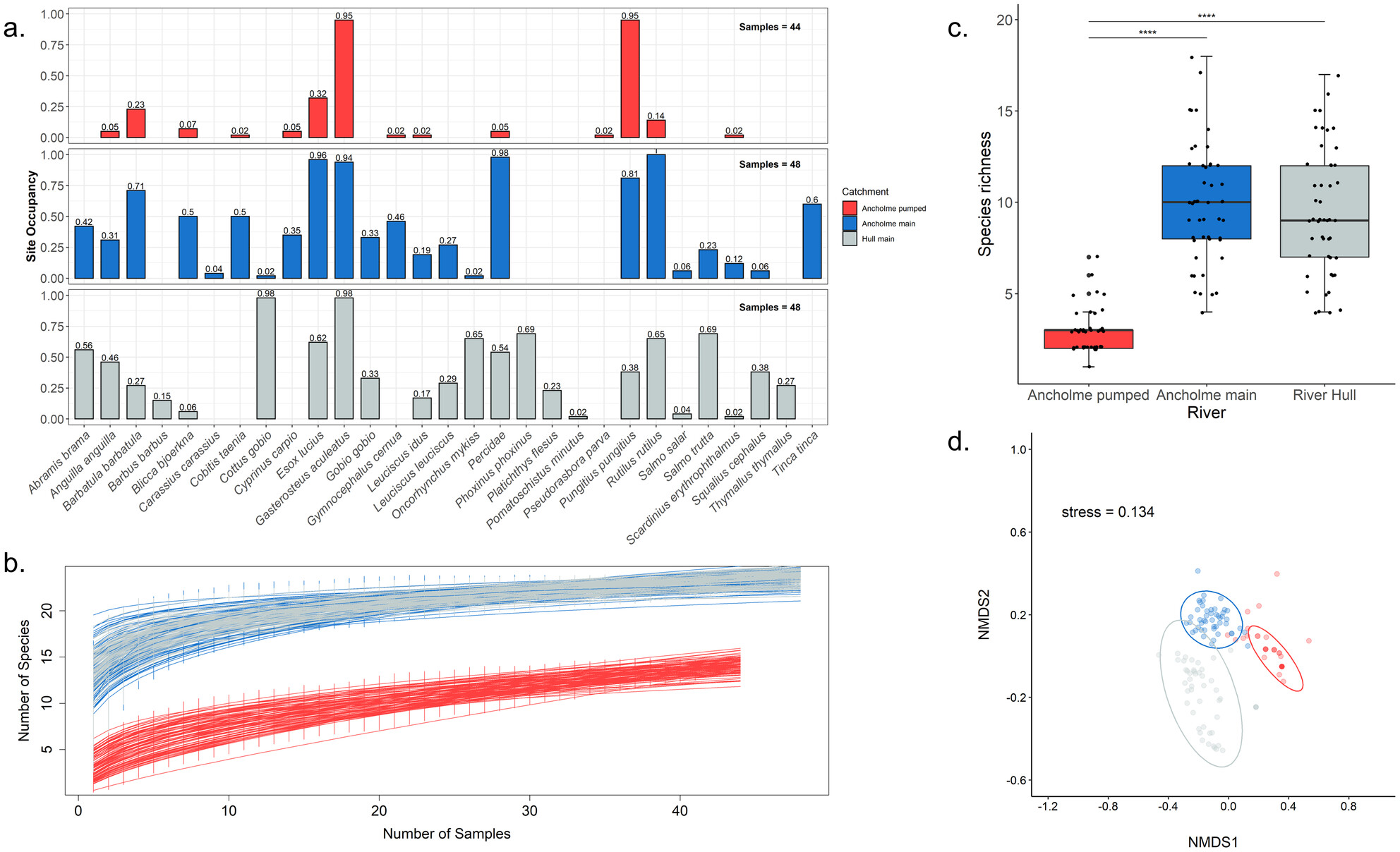
Seasonal Changes in Fish eDNA Signal Vary Between Contrasting River Types
- First Published: 06 June 2025
Warming Increases Environmental DNA (eDNA) Removal Rates in Flowing Waters
- First Published: 03 June 2025
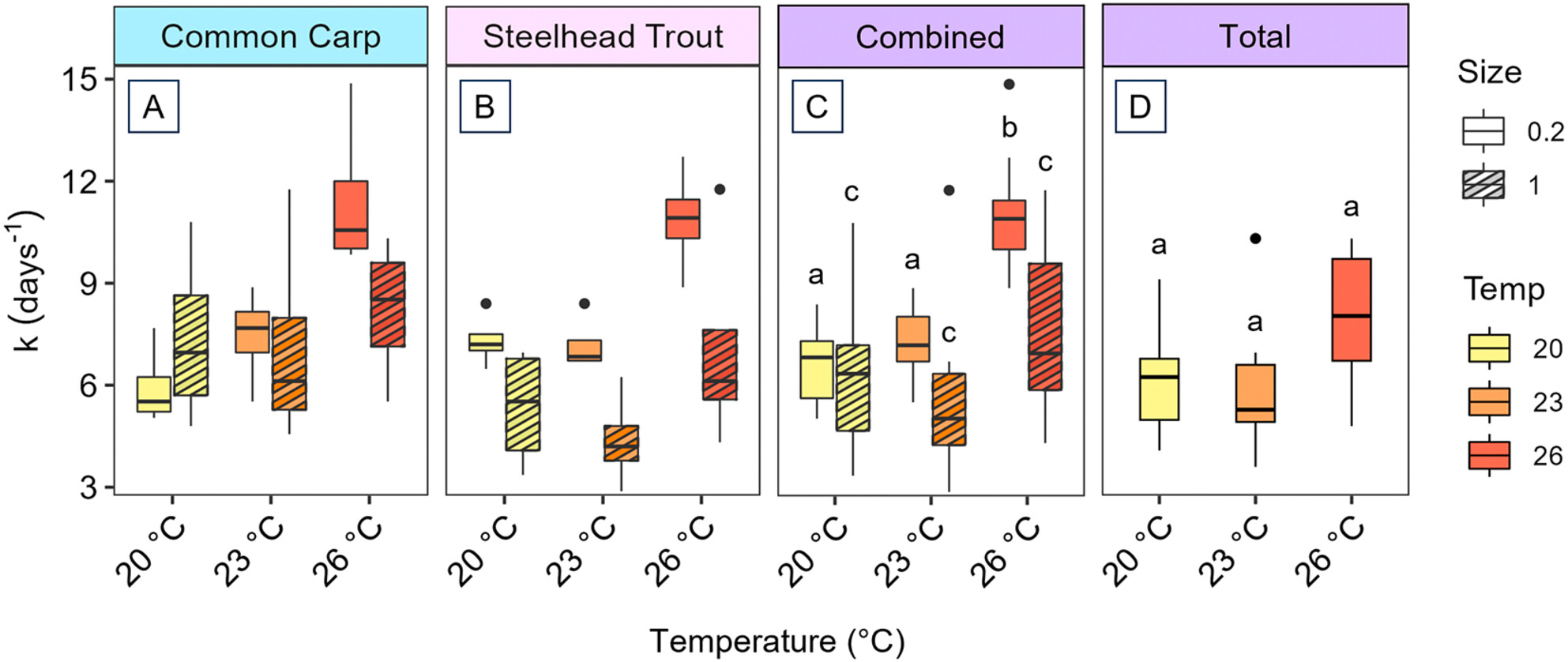
This study experimentally investigated how warmer water temperatures impact eDNA removal rates in flowing waters. Using recirculating mesocosms with varying water temperatures (20°C, 23°C, and 26°C) we found that, for small eDNA particles (0.2–1.0 μm), removal rates were higher at the warmest temperature while removal rates were consistent across temperatures for larger eDNA particles (> 1.0μm). Consequently, smaller eDNA particles were removed faster than larger particles at 26°C and 23°C compared to 20°C, resulting in an increase in the proportion of the eDNA sample made up of small particles with downstream transport for the two warmer temperatures. These results suggest eDNA removal in streams reflects a complex interplay between physical trapping and microbial degradation influenced by temperature, and that differences in temperature between geographic locations, seasons, and climates could impact the fate and interpretation of eDNA in these systems.
Enhancing Environmental DNA Sampling Efficiency for Cetacean Detection on Whale Watching Tours
- First Published: 23 May 2025

Monitoring cetaceans is essential for evaluating ecosystem health and informing the establishment of marine protected areas. This study sought to refine eDNA sampling parameters to offer a more efficient and scalable approach for cetacean research, leveraging citizen science platforms. Our results illustrate that larger water volumes (10 L), immediate sampling post-encounter, and Smith-Root eDNA filters (1.2 μm pore size) significantly increased eDNA detection probability and signal strength.
Quantifying the Temporal Dynamics of Marine Biodiversity Under Anthropogenic Impacts Using eDNA Metabarcoding
- First Published: 26 May 2025
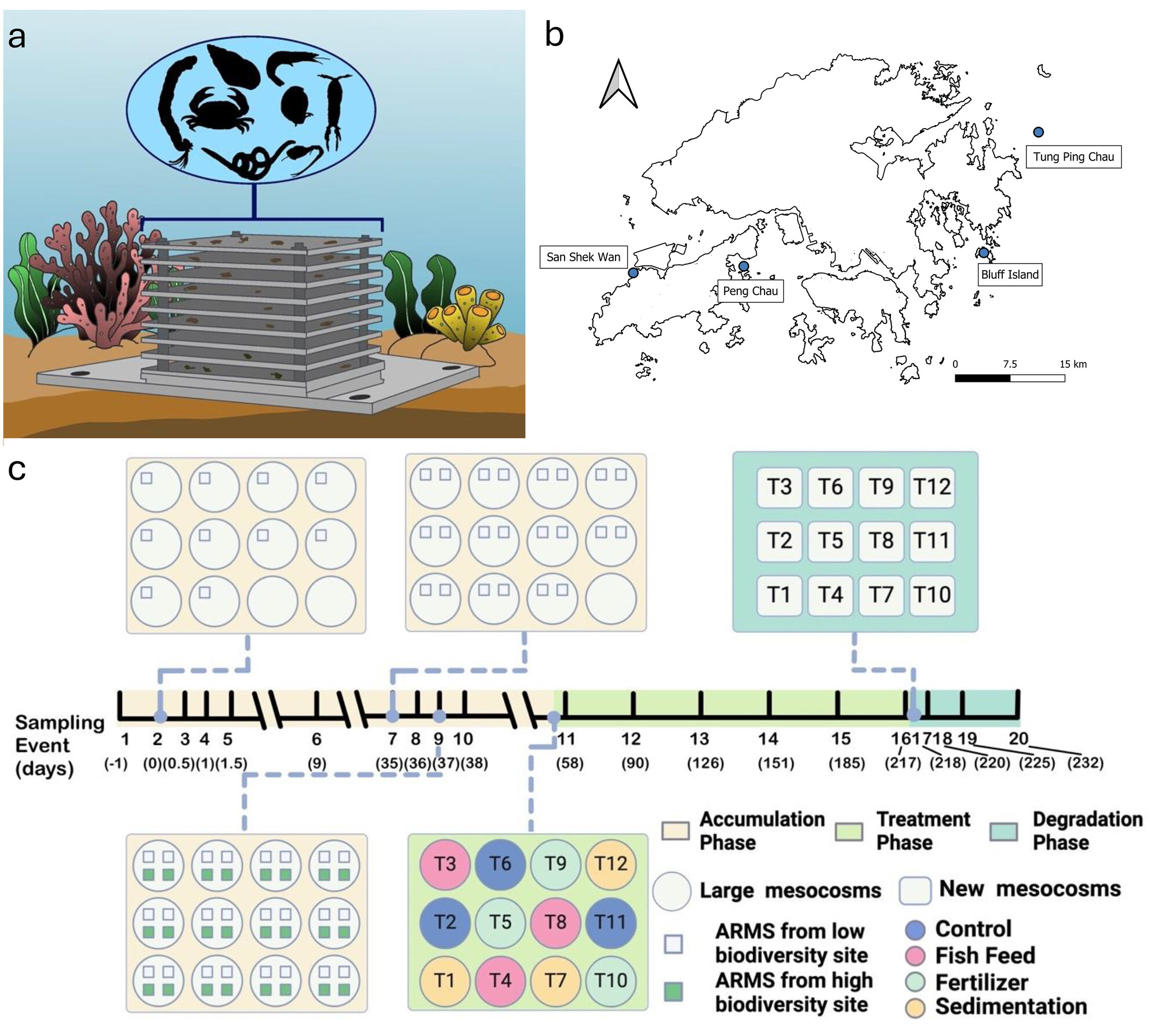
This study uses eDNA to explore marine biodiversity changes under anthropogenic impacts, with a focus on marine community dynamics. Through the use of eDNA metabarcoding, the research identified 814 ASVs across 617 marine families, noting drastic changes in biological communities over time, with different species dominating during different phases of the experiment. The findings highlight the efficacy of eDNA metabarcoding in providing a detailed assessment of biodiversity and differentiating between spatiotemporal and anthropogenic impacts on community dynamics.
Temporal Dynamics of a Fish Parasite (Tetracapsuloides bryosalmonae) and Its Two Main Hosts in Pyrenean Streams: An Environmental DNA-Based Approach
- First Published: 02 June 2025
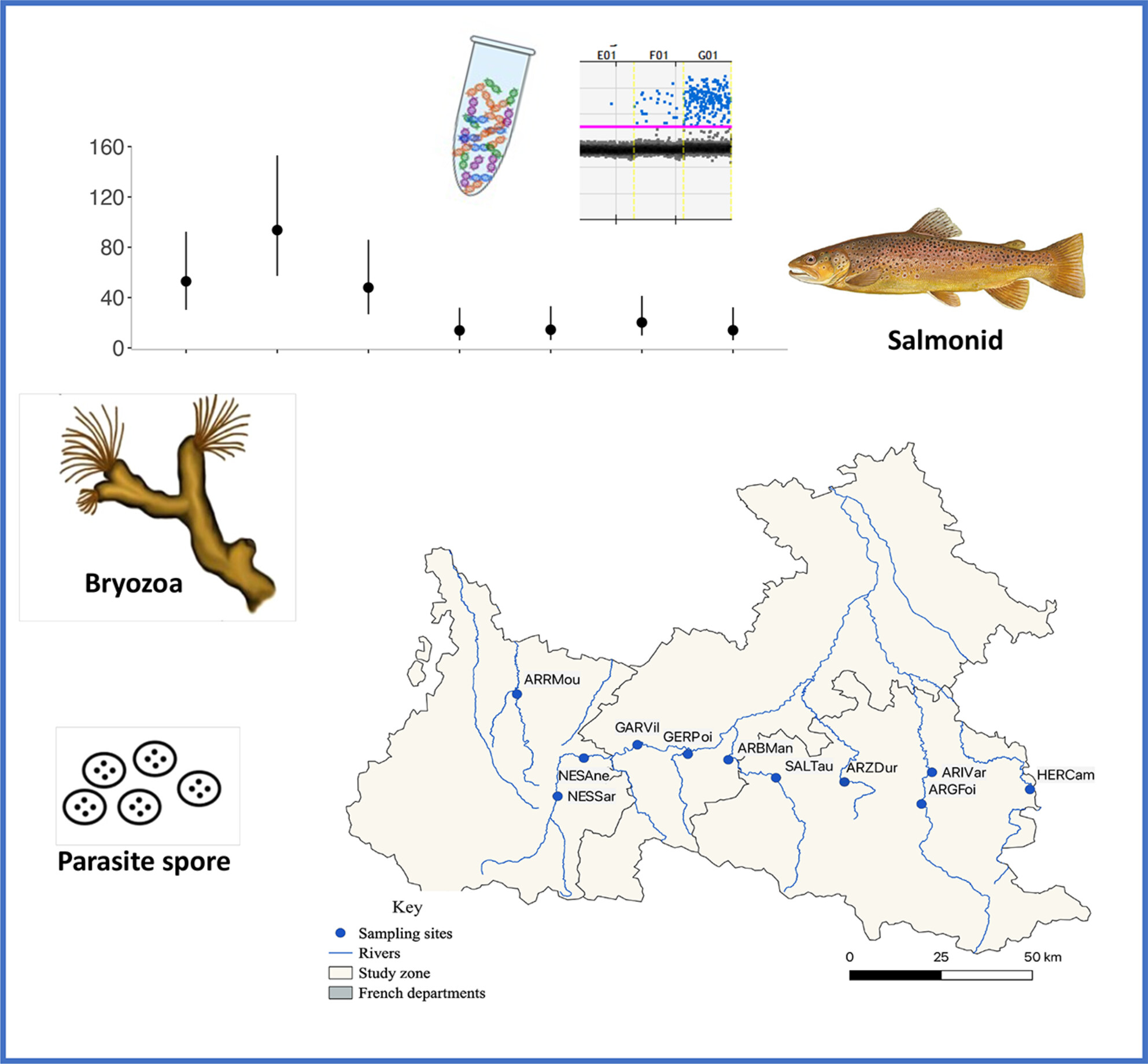
This study used eDNA and digital PCR to track the parasite T. bryosalmonae and its hosts over a year in the Pyrenees. Results showed seasonal and spatial synchrony, especially in summer, with stronger alignment in nearby sites and where fish infection rates were high. The findings suggest synchrony supports the parasite's life cycle and highlight eDNA as an effective tool for studying host-parasite interactions in nature.
Exploring the Dynamics of Environmental DNA: Effects of Early Developmental Stage and Physiological State in Chum Salmon
- First Published: 02 June 2025
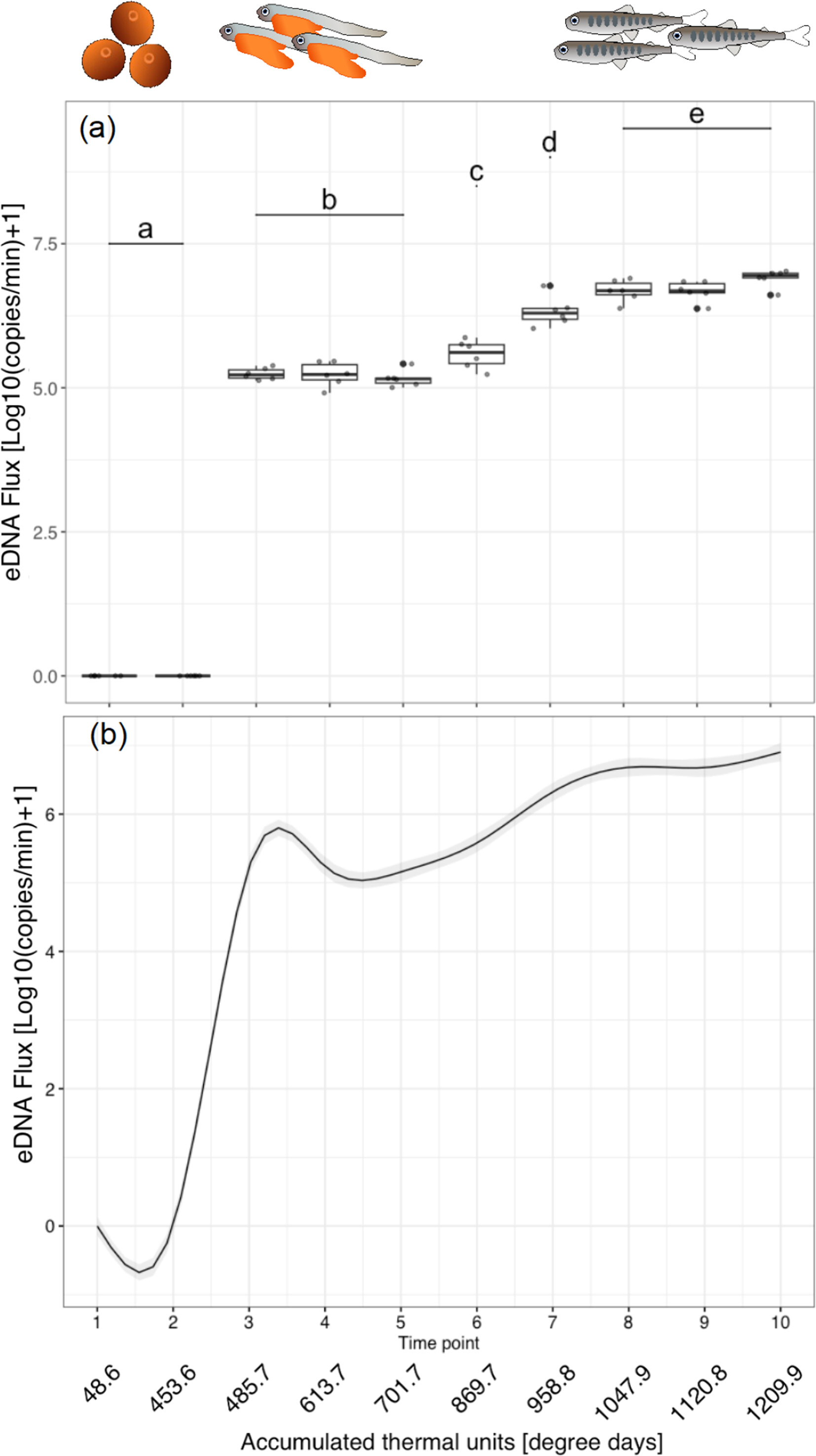
The eDNA flux changes depending on salmon developmental stages. (a) The result of the linear mixed model. The eDNA flux differed significantly among different letters (b-c: p < 0.05, d-e: p < 0.05, Other combinations: p < 0.001). Gray and black dots indicate actual measurements and boxplot outliers, respectively. The eDNA flux at each time point consists of six technical replicates. (b) A generalized additive mixed model (GAMM) fitted for the eDNA flux fluctuation. The eDNA flux at different developmental stages, peaking at Timepoints 8–10 (after the egg yolk is absorbed and floats to the surface and transitions to the juvenile stage). The gray shaded area represents 95% confidence interval.
Gone in a Splash? Temporal Dynamics of Flukeprint Environmental DNA (eDNA) Detection for Common Coastal Northeast Pacific Cetacean Species
- First Published: 30 May 2025
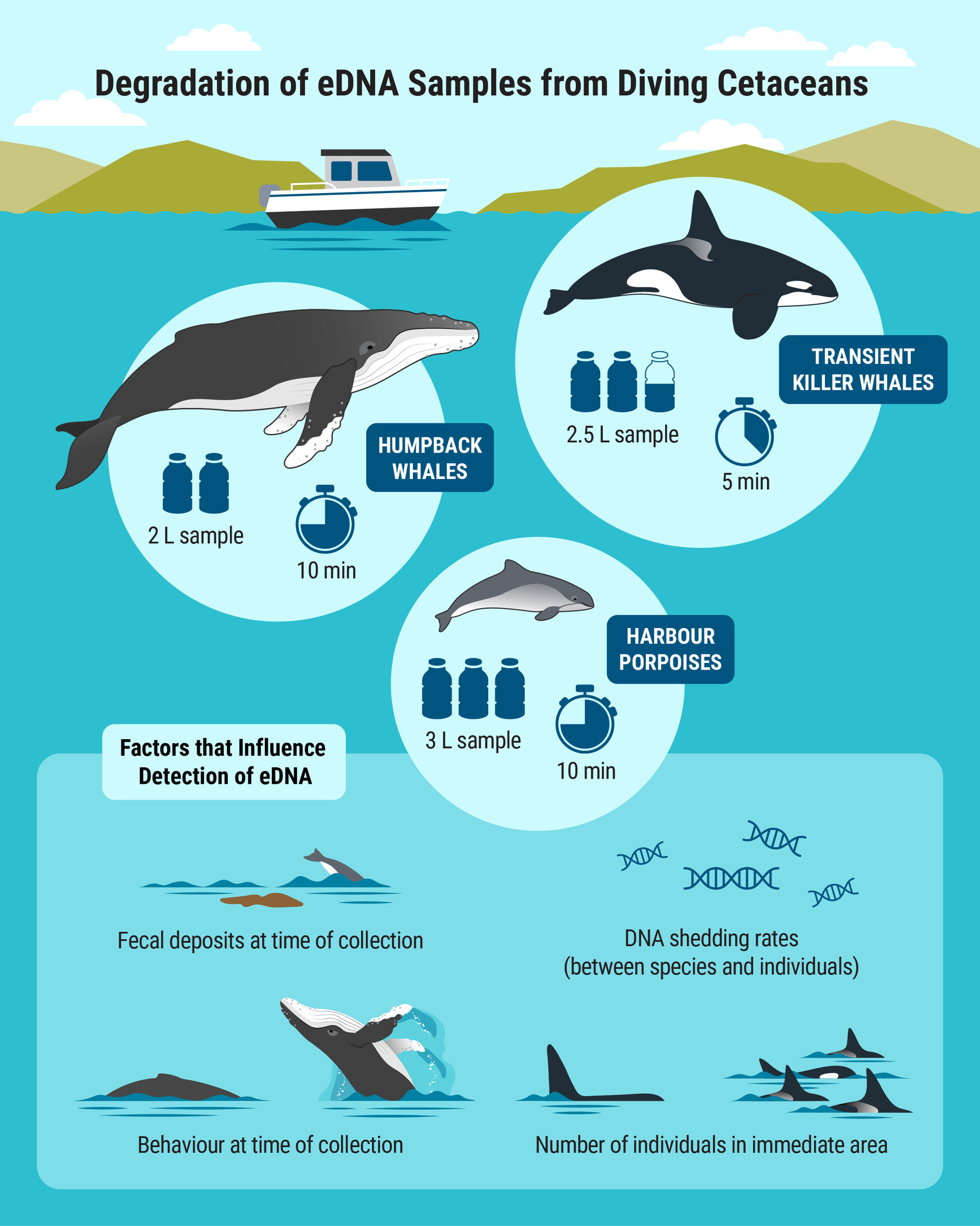
Environmental DNA (eDNA) techniques have been successfully used to detect cetacean species, including harbor porpoises, humpback whales, and killer whales, through direct “flukeprint” sampling. This study assessed the persistence of eDNA shed in flukeprints by collecting seawater samples at timed intervals (up to 10 min) following dives of these species. Results showed variability in eDNA detectability over time, with a general decline in detection, highlighting species-specific differences and the dynamic nature of ocean environments.
Spatial and Seasonal Biodiversity Variation in a Large Mediterranean Lagoon Using Environmental DNA Metabarcoding Through Sponge Tissue Collection
- First Published: 30 May 2025
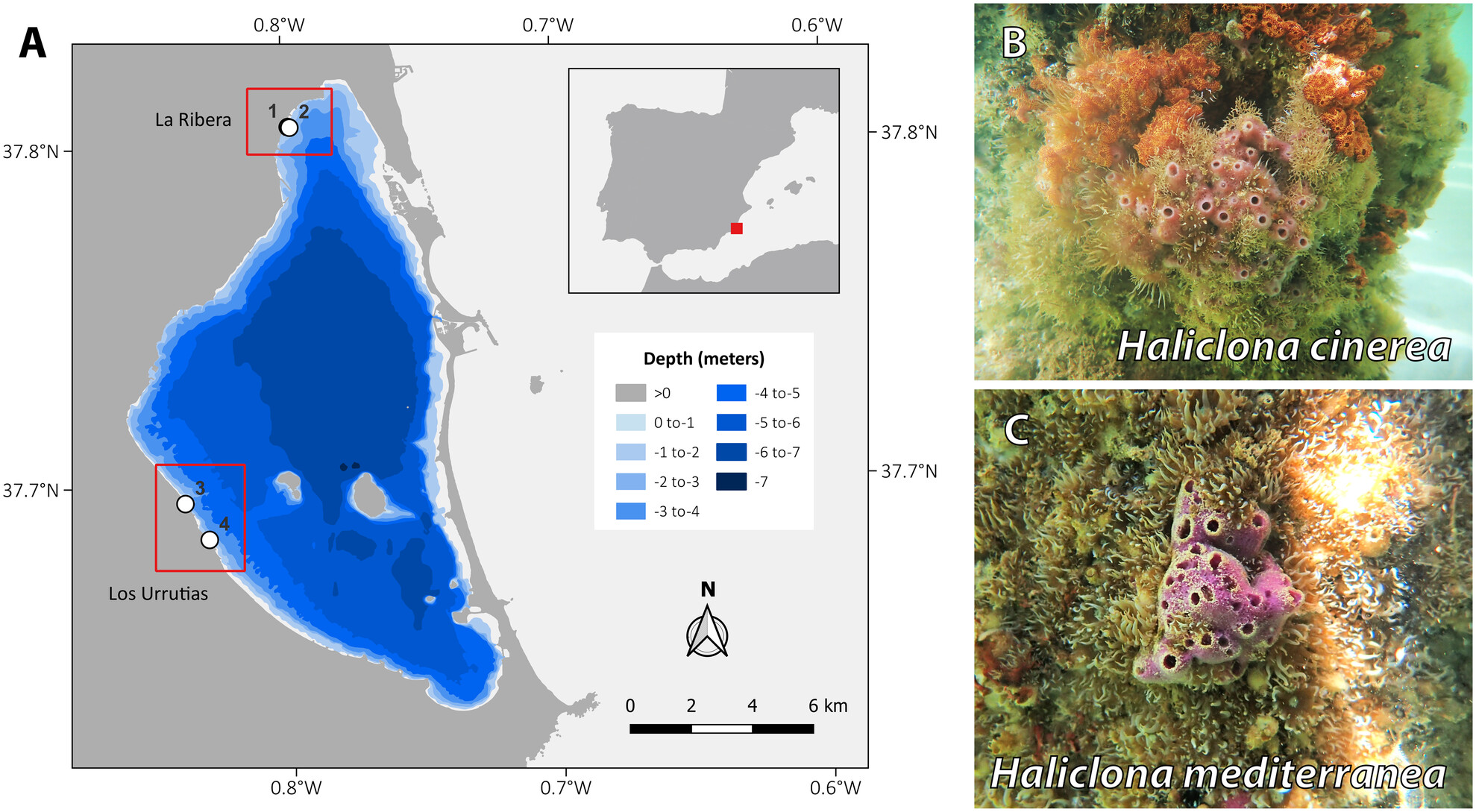
Ecosystem monitoring is essential for preventing biodiversity loss, and in the Mar Menor lagoon (SE Spain), where intensive agriculture has led to eutrophication and mass mortality of marine fauna, it is critical for assessing the status of flora and fauna and informing conservation actions. This study assessed faunal composition by utilizing marine sponges as environmental DNA samplers, identifying 76 taxa, including threatened and invasive species. This cost-effective approach demonstrated high efficacy in characterizing both spatial and temporal biodiversity in shallow coastal ecosystems.
Current Affairs: Examining the Use of Environmental DNA for Relative Abundance Monitoring in a Dynamic Tidal Habitat
- First Published: 28 May 2025
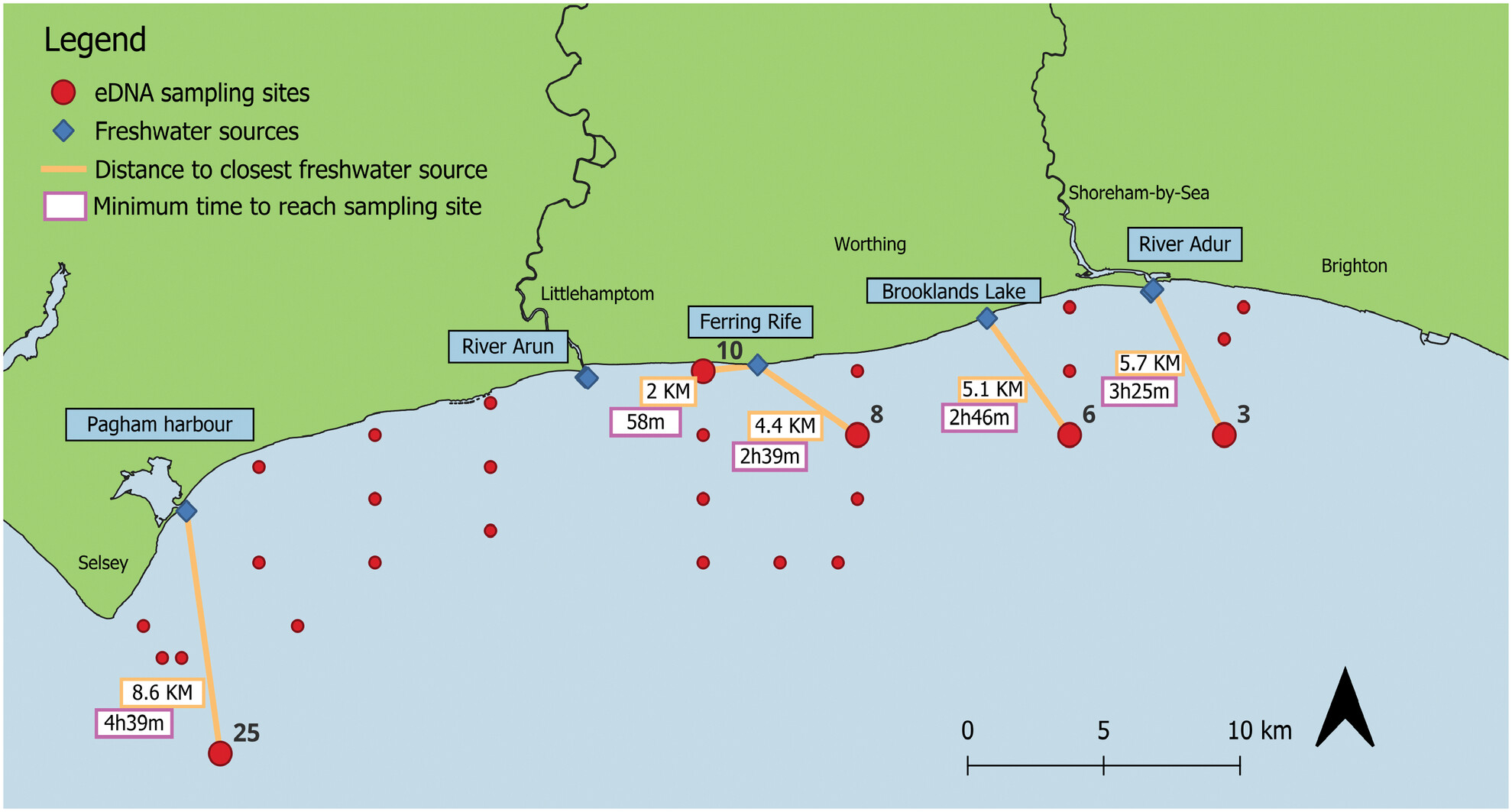
This study evaluates the feasibility of using environmental DNA (eDNA) metabarcoding to estimate the relative abundance of marine species in Sussex Bay, UK. Although eDNA detected more species than Baited Remote Underwater Video (BRUV), a significant correlation between eDNA index and BRUV MaxN was found for only one out of fourteen species analyzed, likely due to differences in detection capabilities and environmental factors. Additionally, we estimated eDNA dispersal in a tidal environment, finding a minimum range of 2–8 km and an average travel speed of 1.8 km/h. Our results highlight the potential of eDNA metabarcoding as a nondestructive biomonitoring tool while also emphasizing its limitations in estimating species abundance in dynamic marine environments.
Release and Degradation of Environmental DNA and RNA From Eels in Aotearoa New Zealand
- First Published: 26 May 2025
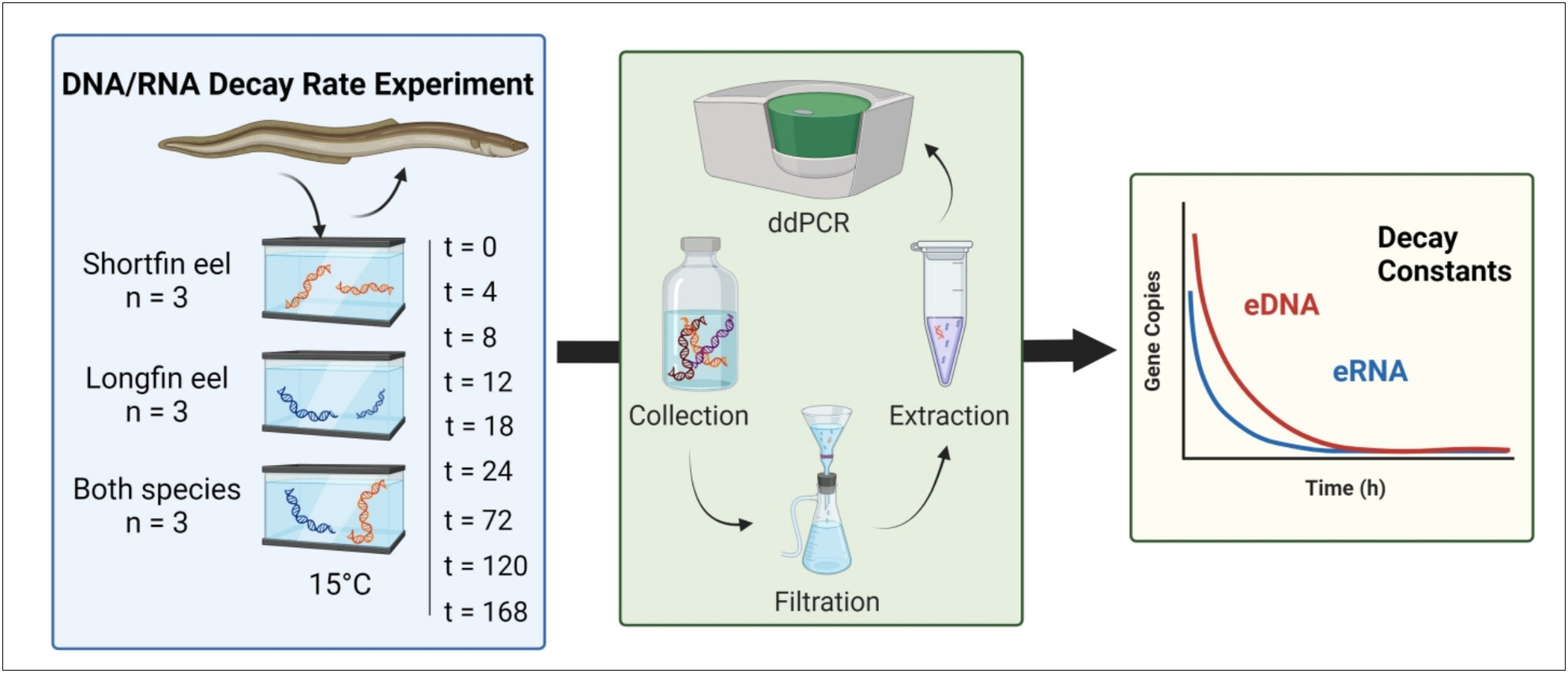
Environmental DNA (eDNA) and RNA (eRNA) dynamics were analyzed in New Zealand longfin and shortfin eels to evaluate their detection potential. Controlled experiments revealed exponential decay patterns, with eRNA degrading faster than eDNA, highlighting its potential for more time-sensitive monitoring. Marker selection and nucleic acid type were key factors influencing detection outcomes.
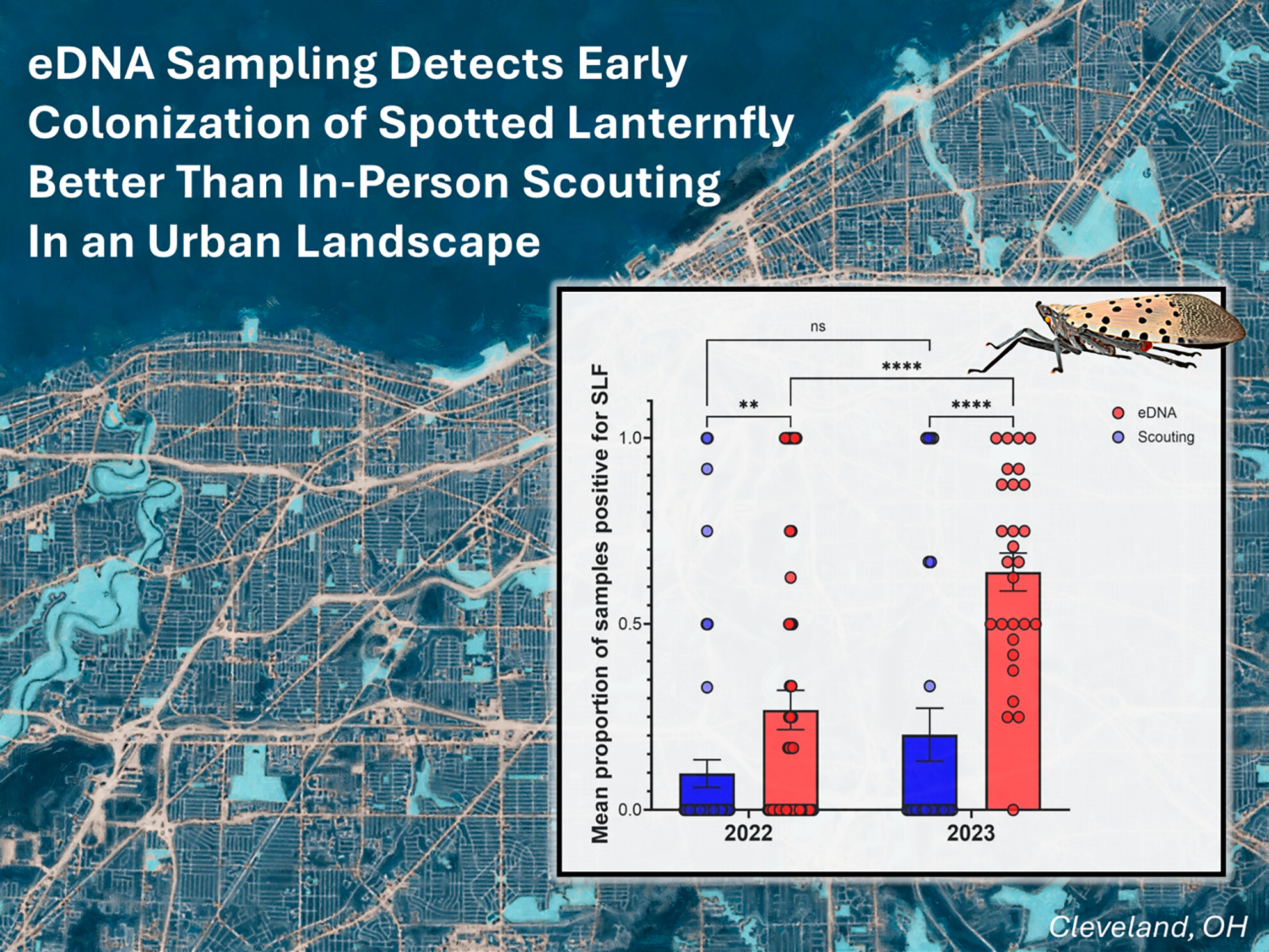
eDNA Sampling Detects Early Colonization of Spotted Lanternfly Lycorma delicatula Better Than In-Person Scouting in an Urban Landscape
- First Published: 26 May 2025
A Time- and Cost-Effective eDNA Protocol to Survey Freshwater Mussels (Bivalvia: Unionida) in Tropical Rivers
- First Published: 08 May 2025
We aimed to develop a reliable and cost-efficient eDNA protocol for surveying tropical freshwater mussels by firstly, developing and validating a qPCR primer-probe assay for Rectidens sumatrensis, and secondly, applying this assay on eDNA collected from lake and river water, respectively, at two different mussel densities in order to test a set of 18 different protocols for capturing, preserving and extracting freshwater mussel eDNA. Mussel eDNA detection and amplification success rates were higher and quantification cycle values were lower for samples preserved in Longmire's buffer compared to ethanol, eDNA captured with filter membrane pore sizes > 0.45 μm, and eDNA extracted with the Qiagen© DNeasy Blood & Tissue Kit rather than the PowerSoil ProKit (albeit latter exhibiting fewer instances of amplification in negative controls).