Exploring the Dynamics of Environmental DNA: Effects of Early Developmental Stage and Physiological State in Chum Salmon
Funding: This work was supported by the Japan Society for the Promotion of Science (17H03623, 23H00329, 26640136).
ABSTRACT
Environmental DNA (eDNA) analyses provide valuable ecological data. Recent studies have explored eDNA dynamics related to reproductive behavior and developmental stages, revealing significant variations in eDNA concentrations across different life stages. However, there is a gap in understanding the association between eDNA concentrations and changes before and after developmental events, such as egg hatching. This study addresses this gap by monitoring eDNA signals in chum salmon (Oncorhynchus keta) during their early developmental stages and examining the effects of their physiological and behavioral changes. For this purpose, eDNA flux was monitored in rearing experiments with chum salmon during their developmental stages (egg, alevin, and fry). The eDNA flux varied significantly across different developmental stages: while no eDNA was detected during the egg stage, eDNA flux increased rapidly after hatching. After hatching, the eDNA flux became stable during the alevin stage but increased approximately 30-fold when they progressed into the fry stage (LMM and post hoc Tukey-HSD test: p < 0.05). These results suggest that eDNA signals vary across the developmental stages and can be utilized to estimate and monitor fish development even under natural conditions, such as those occurring under gravel for salmonid species.
1 Introduction
Environmental DNA (eDNA) analyses, which allow non-invasive and cost-effective investigation of organisms, are used for a variety of purposes such as conservation of rare species, biomass estimation, and biodiversity monitoring (Ballini et al. 2024; Benoit et al. 2023; Wu et al. 2024). The taxonomic groups targeted by eDNA analyses are also diverse, including fish (Timana-Mendoza et al. 2024), amphibians (Mu et al. 2024), reptiles (Nordstrom et al. 2022), mammals (Schenekar et al. 2024), birds (Ushio et al. 2018), invertebrates (Gleason et al. 2021; Marques et al. 2024), and plants (Espinosa Prieto et al. 2023, 2024). The ecology of eDNA, such as degradation (Yamamoto 2024), release (Wood et al. 2020), transport (Jo 2023; Urabe et al. 2024), and state (Brandao-Dias et al. 2023), is also investigated as a basis for application. The origin and source of eDNA have also been discussed (Barnes and Turner 2015; Rodriguez-Ezpeleta et al. 2021). However, analysis of eDNA does not provide information on the size and the number of individuals in the wild (Goldberg et al. 2016). Therefore, the interpretation of eDNA signals depends largely on the inference and the knowledge based on the ecology of the organisms. The dynamics of eDNA signals in response to specific behaviors and changes in developmental stages within the life cycle could provide important insight into the ongoing development of the target species using eDNA.
Few studies have directly examined changes in eDNA concentration across developmental stages during the life cycle of target species. For example, Maruyama et al. (2014) compared eDNA signals from juvenile and adult bluegill sunfish (Lepomis macrochirus). Their results suggested that the eDNA release rate was 3–4 times higher in adults than in juveniles. Takeuchi et al. (2019) compared eDNA concentrations from Japanese eels (Anguilla japonica), but they compared eDNA from different individuals at seven developmental stages. To our knowledge, no studies have been conducted using time-series monitoring to evaluate the changes in eDNA signals from the same individuals. Such monitoring would be essential because the patterns of eDNA signals might be largely affected by the characteristics of individuals, including genetic, metabolic, and behavioral ones (Sassoubre et al. 2016; Wood et al. 2020).
In this study, we conducted rearing experiments with chum salmon (Oncorhynchus keta) to examine temporal changes in eDNA signals throughout their early developmental stages (from egg to fry). Our working hypothesis is a continuous increase in eDNA signals after fertilization, based on the assumption that eDNA release from individuals is closely linked to cell division, irrespective of developmental stages. An alternative hypothesis is a discontinuous increase in eDNA signals, potentially corresponding to physiological and behavioral changes during early development. To test these hypotheses, the specificity of the real-time PCR detection assay was first enhanced to improve the accuracy of eDNA detection from chum salmon. Next, rearing experiments were conducted, and eDNA flux was observed by time-series monitoring, according to physiological and behavioral changes of chum salmon in response to accumulated thermal units (ATUs), as cumulative degree days (Boyd et al. 2010). In this study, ATUs are defined as the cumulative daily mean water temperature (above 0°C). The developmental stage of chum salmon from egg to hatch depends on water temperature, which can be estimated from accumulated experienced water temperatures (Kobayashi 1980; Beacham and Murray 1990). Therefore, we collected water samples at the time points covering egg, alevin, and fry stages based on the ATUs for eDNA analyses (Table 1). Finally, the effects of physiological and behavioral changes associated with the developmental stage on the eDNA signals are examined.
| Timepoint | Sampling date | Days after fertilization | ATUs | Developmental stage | Mean DNA flux (copies/min.) | SD |
|---|---|---|---|---|---|---|
| 0 | 2015.11.13 | 0 | 0.0 | Before setting the eggs | 148.4 | 231.9 |
| 1 | 2015.11.19 | 6 | 48.6 | No eggs hatched | 0 | 0 |
| 2 | 2016.1.8 | 56 | 453.6 | No eggs hatched | 0 | 0 |
| 3 | 2016.1.12 | 60 | 485.7 | 30%–40% of eggs hatched | 178274.4 | 38489.9 |
| 4 | 2016.1.28 | 76 | 613.7 | All eggs hatched | 187269.8 | 76279.0 |
| 5 | 2016.2.8 | 87 | 701.7 | Alevin | 151342.3 | 51449.5 |
| 6 | 2016.2.29 | 108 | 869.7 | Many alevin, some fry | 428648.1 | 199339.5 |
| 7 | 2016.3.11 | 119 | 958.8 | Mostly fry, some alevin | 2475426.7 | 1591714.1 |
| 8 | 2016.3.22 | 130 | 1047.9 | Fry | 5181153.4 | 1877392.3 |
| 9 | 2016.3.31 | 139 | 1120.8 | Fry | 5043850.0 | 1575231.5 |
| 10 | 2016.4.13 | 152 | 1209.9 | Fry | 8355635.6 | 2105719.2 |
2 Materials and Methods
2.1 Modification of Detection Assay for Chum Salmon
Previously developed chum salmon detection assay (Minegishi et al. 2019) turned out to be cross-species amplifiable for O. gorbuscha and Salvelinus namaycush. To improve the usability of this assay in wider environmental conditions, the primer set was modified referring to DNA sequences of the mitochondrial control region and its flanking region of Salmonidae species collected from National Center for Biotechnology Information (NCBI: https://www.ncbi.nlm.nsih.gov) GenBank database as follows: OnKeta-spDL-F5 (5′-TCTGGCGGCTACATCCC-3′), OnKeta-spDL-R5 (5′-GCTGATGTATGAGGGGTTAAAATAAGT-3′). This primer set amplifies 128 bp of the mitochondrial control region of chum salmon, and the previously designed probe (OkDL-Probe1, 5′-[FAM]-CCCATATATAATACTGCACGTGAGTAGTAC-[TAMRA]-3′) is available. To assess the species specificity of this assay, tissue-derived DNA from ten salmonid fish species co-existing in Japan was subjected to real-time PCR analyses: O. keta, O. nerka, O. masou masou, O. gorbuscha, O. mykiss, and Salv. leucomaenis, Salv. curilus, Salv. namaycush, Salmo trutta, and Parahucho perryi. Optimized real-time PCR conditions and settings were used for this in vitro test and subsequent validation analyses. The details were described in the “eDNA analysis” section. In addition, as an in silico PCR, Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome) was performed to search for potentially amplifiable species by the modified primer pair in all fish sequences registered at NCBI. Primer-BLAST was performed on fish sequences with the database set to “nr” and other settings set to default. The limit of detection (LOD) and the limit of quantification (LOQ) of this assay were assessed using a 10-fold dilution series of standards (2 to 2 × 105 copies/reaction) with ten technical replicates. The definition of LOD and LOQ was the lowest standard concentration that could be amplified in 95% of replicates, and the lowest standard concentration with a coefficient of variation < 35%, respectively (Klymus et al. 2020). The LOD and LOQ were calculated using the LOD/LOQ calculator R script (Klymus et al. 2020). For further validation, a qPCR product from each tank experiment (the next section) was sequenced in both directions using BigDye Terminator v1.1 Cycle Sequencing kit (Thermo Fisher Scientific, Waltham, MA) on a SeqStudio Genetic Analyzer (Thermo Fisher Scientific).
2.2 Experimental Set-Up and Water Sampling
We conducted rearing experiments to verify that the fluctuation of the eDNA signal depends on the developmental stage. The experiment was conducted at the Chitose Salmon Hatchery Station of the Fisheries Resources Institute, Japan Fisheries Research and Education Agency (Figure 1). We prepared three flow-through tanks, called modified Atkins incubators, which held 101 L (146 × 33 × 21 cm) of water during the experiment. Spring water was used as inlet water into each tank (7 L/min). We set 2500 chum salmon eggs in each tank (wet weight, 516.2, 515.0 g, and 515.4 g). The water temperature was recorded daily and remained stable between 8.0°C and 8.1°C throughout the experiment (Table S1). The rearing experiment was conducted from before egg introduction (Timepoint = 0) to the time point when the accumulated water temperature was 1209.9ATUs (Timepoint = 10), from Nov. 13, 2015, to Apr. 13, 2016, and a total of 11 water samples were performed (Table 1). Pellet feeding was started on Mar. 2nd, 2016 (between Timepoints 6 and 7), at a level of 3% of estimated total individual wet weight. Salmon biomass (g) in each tank was measured at or around the time points 0 (Nov. 13th, 2015), 6 (Mar. 1st, 2016), and 8 (Mar. 24th, 2016) as the wet weight of all individuals in each tank. We collected a 1 L water sample from the inlet (tank negative control) and two 1 L water samples from the outlet of each tank with 1 L disposable plastic cups. In total, we collected 66 tank water samples (2-sampling replicates × 3 tanks × 11 Timepoints) and 33 inlet water samples. Immediately after the sampling, we filtered the water samples with glass-membrane filters with a mesh size of 0.7 μm (Whatman GF/F, GE Healthcare Japan, Tokyo, Japan). As the final step of filtering, we filtered 10 mL of 70% ethanol to fix the samples. All filter samples were stored at −25°C until further analyses. To prevent cross-contamination, all filtration equipment was sterilized and carefully rinsed with pure water after each filtration.

2.3 eDNA Analysis
2.4 Statistical Analysis
A generalized additive mixture model (GAMM) with a Gaussian distribution was used to examine changes in eDNA signals at different developmental stages. This analysis was performed using the “gamm” function in the “mgcv” package (version 1.9–1) in R (version 4.3.3). In this model, log-transformed eDNA flux was used as the response variable. The time point (developmental stage) was set as an explanatory variable, and Tank ID was set as a random effect. To compare the change in eDNA flux owing to the difference in developmental stages, a linear mixed model (LMM) and a post hoc Tukey–HSD test were performed, using the “glht” function in the “multcomp” package (1.4–25). This model used log-transformed eDNA flux as the response variable, the time point (developmental stage) as an explanatory variable, and the Tank ID as a random effect. Another LMM and a post hoc Tukey–HSD test were conducted to compare eDNA flux per biomass of salmon across different developmental stages. The log-transformed eDNA flux per salmon biomass was used as the response variable, with Timepoint as an explanatory variable and Tank ID as a random effect. All graphs were drawn using the “ggplot2” package (version 3.4.2).
3 Results
For evaluating the modified chum salmon eDNA detection assay, an in vitro test was performed using the tissue-derived DNA from ten salmonid species. The results confirmed that the assay amplified only DNA from chum salmon (Figure S1). In addition, the result of in silico PCR (Primer-BLAST to bony fish database) indicated that the modified primer pair amplified only the target species among bony fishes. The LOD was 8.64 copies/reaction, and the LOQ was 29 copies/reaction. The DNA sequences from qPCR products obtained from water samples in the three tanks at Timepoint 8 in the rearing experiment were then analyzed using Sanger sequencing and confirmed to perfectly match the chum salmon DNA sequence available in Genbank (e.g., LC616475.1).
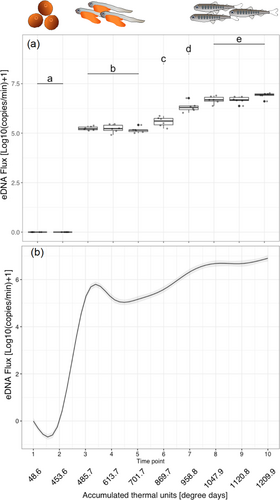
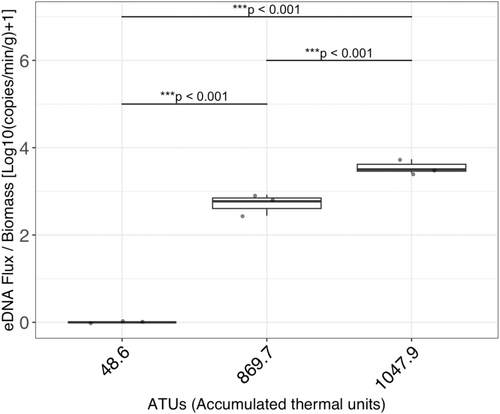
The R2 values of calibration curves were > 0.995 in all runs. The slopes ranged between −3.766 and −3.365, and the PCR efficiency ranged between 84.3% and 98.2%. The DNA flux at each time point is shown in Table 1. Results of the rearing experiments showed that the eDNA flux varied with the developmental stages (GAMM: p < 0.001). Measured salmon biomass was 516.2, 515.0, and 515.4 g at Timepoint 1 (ATUs: 48.6, Average ± SD: 515.5 ± 0.50); 756.8, 752.2, and 753.7 g at Timepoint 6 (ATUs: 869.7, Average ± SD: 754.2 ± 1.92); and 1381.4, 1374.1, and 1375.2 g at Timepoint 8 (ATUs: 1047.9, Average ± SD: 1376.9 ± 3.21) (in Tank 1, 2, and 3, respectively). The results of eDNA quantification and smoothing by GAMM are shown in Figure 2a,b, respectively. In particular, no eDNA detection was observed in the egg period (Timepoints 1 and 2). After hatching occurred (between Timepoints 2 and 3), the eDNA flux increased rapidly. The eDNA flux was stable during the alevin stage (Timepoints 3, 4, and 5), but it increased on average 30.3-fold between Timepoints 3–5 and 8 along with the transition from alevin to fry stage. The eDNA flux was significantly different among groups a (Timepoints 1 and 2), b (Timepoints 3, 4, and 5), c (Timepoint 6), d (Timepoint 7), and e (Timepoints 8, 9, and 10) in Figure 2a (LMM and post hoc Turkey-HSD test: p < 0.05). In addition, DNA flux per salmon biomass (g) differed significantly across developmental stages (among egg, alevin, and fry, all p < 0.001, Figure 3).
4 Discussion
In this study, we elucidated the dynamics of eDNA flux based on the monitoring of the eDNA flux from the same individuals of chum salmon from the egg to fry stages over time in the rearing experiment. No eDNA was detected during the egg stage, but a sharp increase in eDNA flux was observed after hatching. In addition, there were significant changes in eDNA flux during the transition from alevin to fry stages. These results clearly contradicted the working hypothesis of a continuous increase in eDNA release from individuals after fertilization. Furthermore, the amount of eDNA released was not solely determined by their biomass and varied significantly across developmental stages (Figure 3). Thus, our results support the alternative hypothesis that the eDNA release increases discontinuously during the developmental stage transitions. These findings are consistent with a previous study in which eDNA concentrations at different developmental stages were examined (Takeuchi et al. 2019). Given that our experiments tracked the same individuals throughout the measurements, the alternative hypothesis is supported rather strongly in this study because individual variations cannot explain the observed discontinuity. These findings are crucial as a foundational understanding of eDNA from fish species because the effects of life-stage events and associated changes in physiological conditions were clarified in this study.
The eDNA signal varied depending on the developmental stage. The developmental stage of chum salmon, from egg to fry, is regulated by water temperature and can be estimated based on the cumulative water temperature experienced (Kobayashi 1980; Beacham and Murray 1990). Being consistent with the previous report (chum salmon eggs hatch at an accumulated water temperature of 480.0ATUs, Kobayashi 1980), eggs did not hatch until Timepoint 2 (453.6ATUs) and hatched at Timepoint 3 (485.7ATUs) in this experiment. After the transition, eDNA became detectable at a high level (133886.7–241931.7 copies/min). Little eDNA may be released from the egg membrane, but it is possible that components within the egg were released upon hatching, becoming a significant source of eDNA. When the accumulated water temperature reaches around 900°C to 1000°C (ATUs in this study), the yolk is expected to be absorbed (Salo 1991), and the individuals start to swim (Crisp 2000). At that time, a physiological transition occurs from yolk-derived endotrophs to exotrophs, and the fish begin to emerge from redd to swim and start retrieving food (Crisp 2000). Prior to floating, there is minimal activity, such as feeding and excretion, with nutrients being absorbed from the yolk. Thus, it is understandable that the eDNA flux remained constant during Timepoints 3, 4, and 5 (485.7, 613.7, and 701.7ATUs, respectively). With the increased number of emerging individuals, the eDNA flux also increased during Timepoints 6, 7, and 8 (896.7, 958.8, and 1047.9ATUs, respectively). Interestingly, however, the eDNA flux was stabilized after Timepoint 8. These results suggest that the physiological status significantly influences the release of eDNA per individual and that the eDNA release per biomass differs among developmental stages, as shown in Figure 3. While an adult stage was not included in this study, there may be no discontinuity between juvenile and adult stages according to previous studies (Mizumoto et al. 2018; Karlsson et al. 2022, but also see Maruyama et al. 2014). It is also noteworthy that juvenile and adult stages tend to share similar physiological traits, relative to the early developmental stages studied in this study (egg to fry). Integrating these findings with those of the present study, it is likely that changes other than biomass associated with changes in developmental stage, that is, changes in physiological state, cause the rapid and discontinuous increase in eDNA signaling. Although this study focused on changes in eDNA signals in rearing experiments during periods when field observations are challenging, the rapid shifts in eDNA signals might also be detectable in natural environments as an indicator of favorable growth in salmon at their early developmental stage, which occurs underground.
It is also likely that the excretion associated with feeding behavior, such as feces, is one of the largest sources of eDNA in the water. Aquatic organisms such as fish and other vertebrates are considered to release DNA as epidermis, mucus, scales, feces, and gametes (Barnes and Turner 2015; Rodriguez-Ezpeleta et al. 2021). The rapid increase in eDNA flux observed after the transition of developmental stages does not necessarily define the origin of eDNA. However, it suggests that egg contents and feces may constitute major eDNA sources. Given that salmon alevin obtains energy solely from the yolk sac and that feces can be an eDNA source only after the fry stage, eDNA from feces might be a part of the causes for the discontinuous increase in eDNA signal between the alevin and the fry stages.
In fish, eDNA concentrations are also known to spike during reproductive behavior (Bracken et al. 2019; Di Muri et al. 2022; Wu et al. 2022). It is also known that the nuclear DNA/mitochondrial DNA ratio increases during their reproduction, mainly due to the release of large numbers of sperm with a low percentage of mitochondrial DNA compared to somatic cells (Bylemans et al. 2017; Wu et al. 2023). In fact, some attempts have been made to estimate reproductive behavior using changes in eDNA concentration and nuclear DNA/mitochondrial DNA ratios (Di Muri et al. 2022; Wu et al. 2023). Therefore, combining the existing knowledge and techniques with the findings in this study, it will be possible that periodic eDNA monitoring will show the following eDNA fluctuations in organisms with synchronized reproductive behavior: (1) a spike in eDNA concentration (and change in nuclear DNA/mitochondrial DNA ratio) due to reproductive behavior; (2) almost no detection of eDNA at the egg stage; (3) sharp increase of eDNA concentration after hatching (alevin phase); (4) further increase of the eDNA concentration after transition to fry (i.e., the egg yolk is absorbed and fish becomes an exotroph). This fluctuation will allow more accurate monitoring of the progression of the life cycle of the target species.
In a previous assay, there was a risk of inadvertently detecting non-target species, particularly in places hosting multiple salmonid species (e.g., Hokkaido, Japan. Imamura et al. 2020; Kanbe et al. 2023). In this study, the issue regarding chum salmon eDNA was addressed with the modified detection assay. Field surveys utilizing the eDNA of chum salmon, which represent a significant biomass among the salmonid species in Japan and other countries hosting this species, can be conducted with high accuracy (see Minegishi et al. 2019). In the case of semelparous organisms such as chum salmon, adult fish die after spawning. Therefore, the potential source of chum salmon eDNA after their spawning season would be the next generation, unless salmon carcasses remain in the spawning grounds. This is ideal for eDNA monitoring of fish reproduction in the wild, and our results suggest that there is a potential for eDNA that enables non-invasive observation of the healthy growth of the fish under gravel and their biomass estimation.
In conclusion, time-series monitoring in rearing experiments involving developmental stage transitions suggested that little eDNA is released from eggs, but its concentration sharply increases upon hatching. Additionally, physiological changes associated with developmental stage transitions were also reflected in eDNA concentration. Particularly, significant changes in eDNA release were observed during the transition from the larval stage, characterized by the absorption of the yolk sac and no feeding or excretion, to the juvenile stage. It is expected that such changes in eDNA signals can be observed in natural river environments as well, although further studies are necessary to confirm this. By detecting eDNA signals in the field, it may be possible to non-invasively monitor the healthy growth of the population. In addition, distinguishing the developmental stages of target species has recently been attempted using environmental RNA (eRNA) (Parsley and Goldberg 2023), but it is still in underdevelopment. Clarification of the dynamics of eDNA signals during the transition of developmental stages will further develop and extend the scope of interpretation of applications already underway, such as eDNA monitoring in the field.
Author Contributions
H.A. conceived and designed the study. H.A. and S.S. collected samples, and T.K. performed laboratory experiments. M.K.S. performed the statistical analysis and wrote the first draft of the manuscript. All authors discussed the results and contributed to the development of the manuscript.
Acknowledgments
The experiments were conducted in the Chitose Salmon Hatchery Station, Salmon Enhancement Division, Salmon Research Department, Fisheries Resources Institute, Japan Fisheries Research and Education Agency (FRA) in the city of Chitose, Japan. We thank Manabu Kagaya from the Chitose Salmon Hatchery Station for his help with experimental set-up and rearing experiment, and Tomoki Sato from the Salmon Resources and Ecology Division, Salmon Research Department, Fisheries Resources Institute, FRA, for his technical advice and help with water sample filtering. We also thank members of the Animal Ecology Lab in the Faculty of Agriculture, Hokkaido University, for technical help. This study was funded by JSPS KAKENHI grant numbers 26640136, 17H03623, and 23H00329 (H.A.).
Ethics Statement
This study was conducted following the ARRIVE guidelines (Japan), and it complies with the International Council for Laboratory Animal Science (ICLAS) guidelines for the treatment of animals in research.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
All raw data are included in the Supporting Information: Figure S1 and Table S1.




