Journal list menu
Export Citations
Download PDFs
ISSUE INFORMATION
ORIGINAL ARTICLE
Invasive Crayfish: Drivers or Passengers of Degradation in Freshwater Ecosystems?
- First Published: 02 March 2025
METHOD
A Confidence Scoring Procedure for eDNA Metabarcoding Records and Its Application to a Global Marine Fish Dataset
- First Published: 20 March 2025
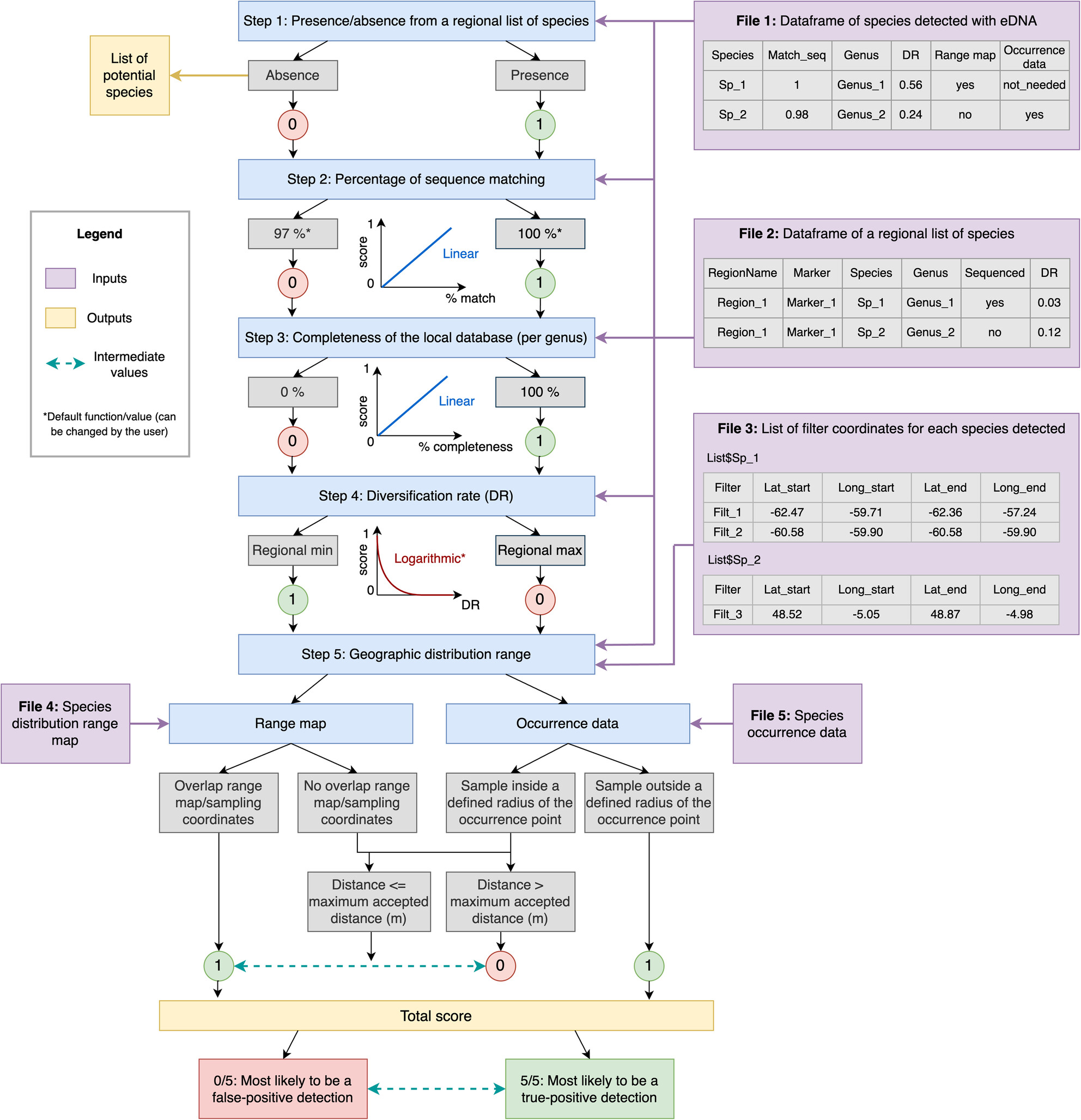
Metabarcoding of environmental DNA (eDNA) is transforming biodiversity detection and surveys across ecosystems and is becoming part of biodiversity monitoring programs. However, interpretations of eDNA records vary widely, from treating them as standard biodiversity records to requiring careful validation against taxonomic and biogeographic data. A new five-step procedure assesses the confidence of species-level eDNA records, revealing that confidence varies by region due to incomplete reference databases and taxonomic resolution issues.
ORIGINAL ARTICLE
Validated Environmental DNA Assay for Detection of the Rare Rice's Whale (Balaenoptera ricei)
- First Published: 20 March 2025
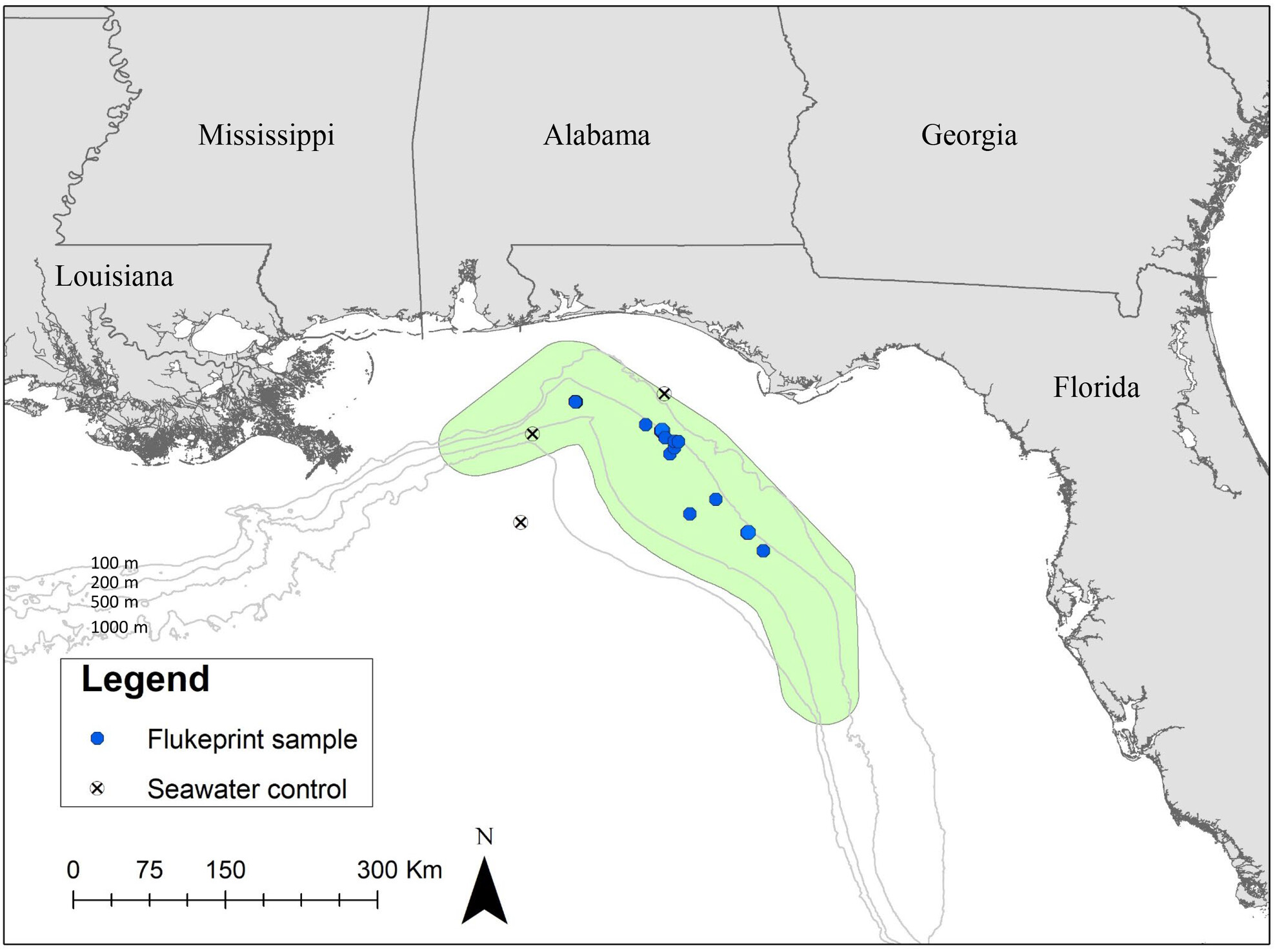
The first eDNA assay was developed to detect the presence of the Rice's whale from environmental samples. This validated qPCR assay is highly sensitive and species-specific and can be utilized in future studies to better understand the temporal and spatial distribution of this rare and endangered species.
Overwintering Fish Community in Ice-Covered Environments in Hokkaido, Japan, Inferred From Environmental DNA
- First Published: 04 March 2025
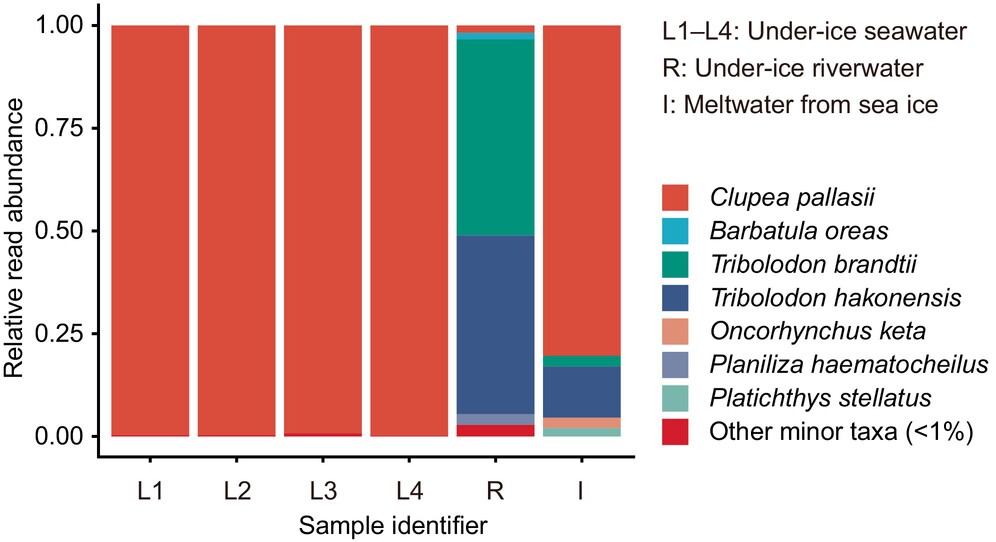
Using fish eDNA metabarcoding, we successfully described an overwintering fish community in ice-covered environments. eDNA data indicated that the dominant species were Clupea pallasii in the lagoon and Tribolodon spp. in the inlet river. Our findings suggested that the eDNA technique is useful for studying ice-covered areas and could provide new insights into the overwintering ecology of fish.
Comparing the Fate of eDNA by Particle Sizes and Molecule Lengths in Recirculating Streams
- First Published: 04 March 2025
Real-Time PCR Assay and Environmental DNA Workflow for Detecting Irukandji Jellyfish, Malo bella (Cubozoa)
- First Published: 06 March 2025
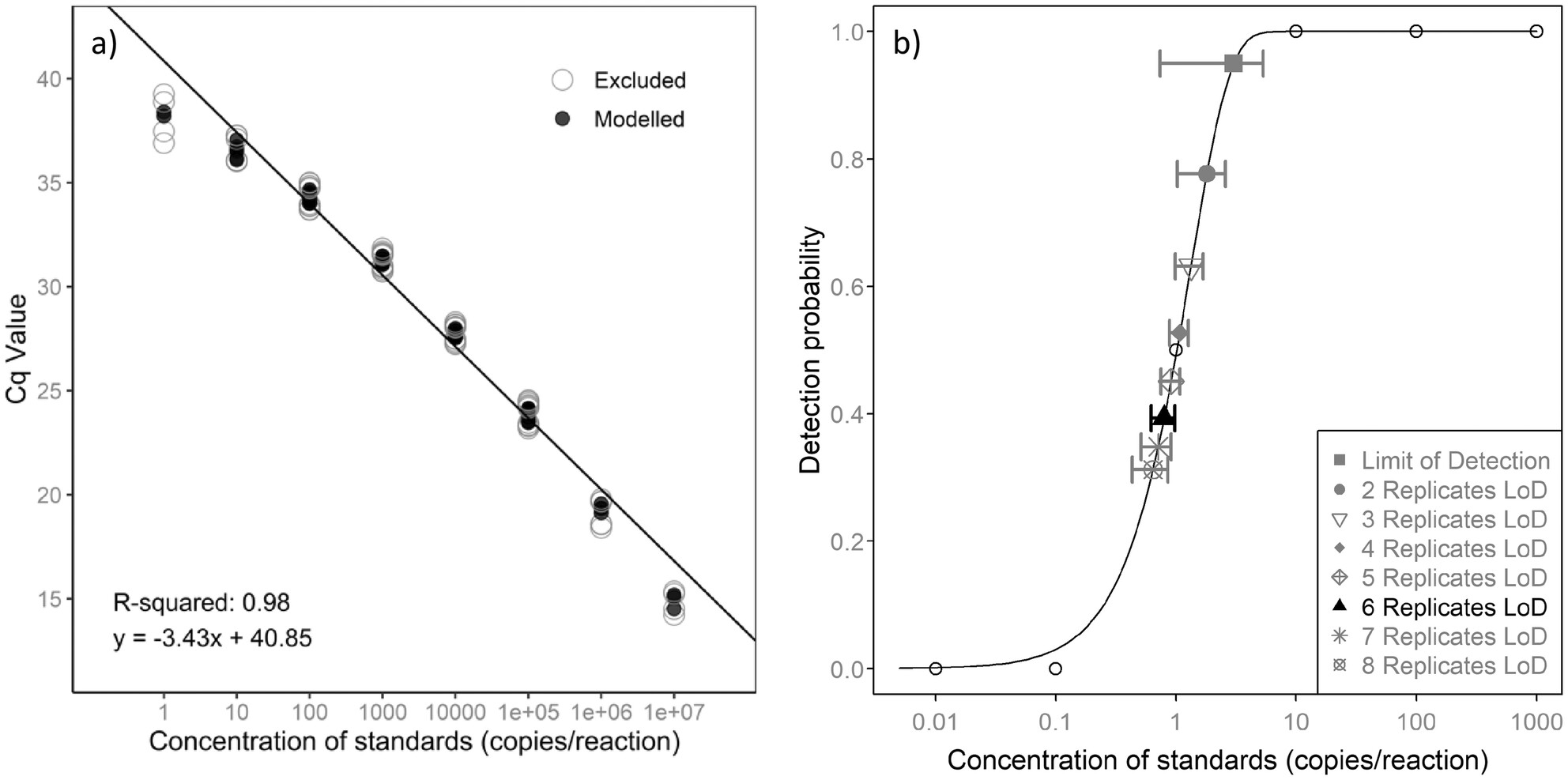
This paper details the development and validation of a highly sensitive and specific quantitative PCR assay to detect and monitor Malo bella, a venomous Irukandji jellyfish species. Using targeted primers and a TaqMan MGB probe, the assay effectively detects M. bella environmental DNA in water samples. This method addresses crucial knowledge gaps in the species' ecology, particularly its spatial and temporal distributions, and enhances the management of envenomation risks in tourism hotspots.
There is no Smoke Without Fire: Evaluation of Water eDNA Profile of Yersinia ruckeri to Assess Enteric Redmouth Disease Pathogenesis in Rainbow Trout (Oncorhynchus mykiss)
- First Published: 06 March 2025
REVIEW
eDNA Metabarcoding Applications Across Italian Marine Coastal Ecosystems: An Overview
- First Published: 06 March 2025
ORIGINAL ARTICLE
Temporal and Spatial Dynamics of the Lotic Fish Communities: A Comparison of Coffee Filter-Based Passive eDNA Collection Versus Active eDNA Filtering
- First Published: 06 March 2025
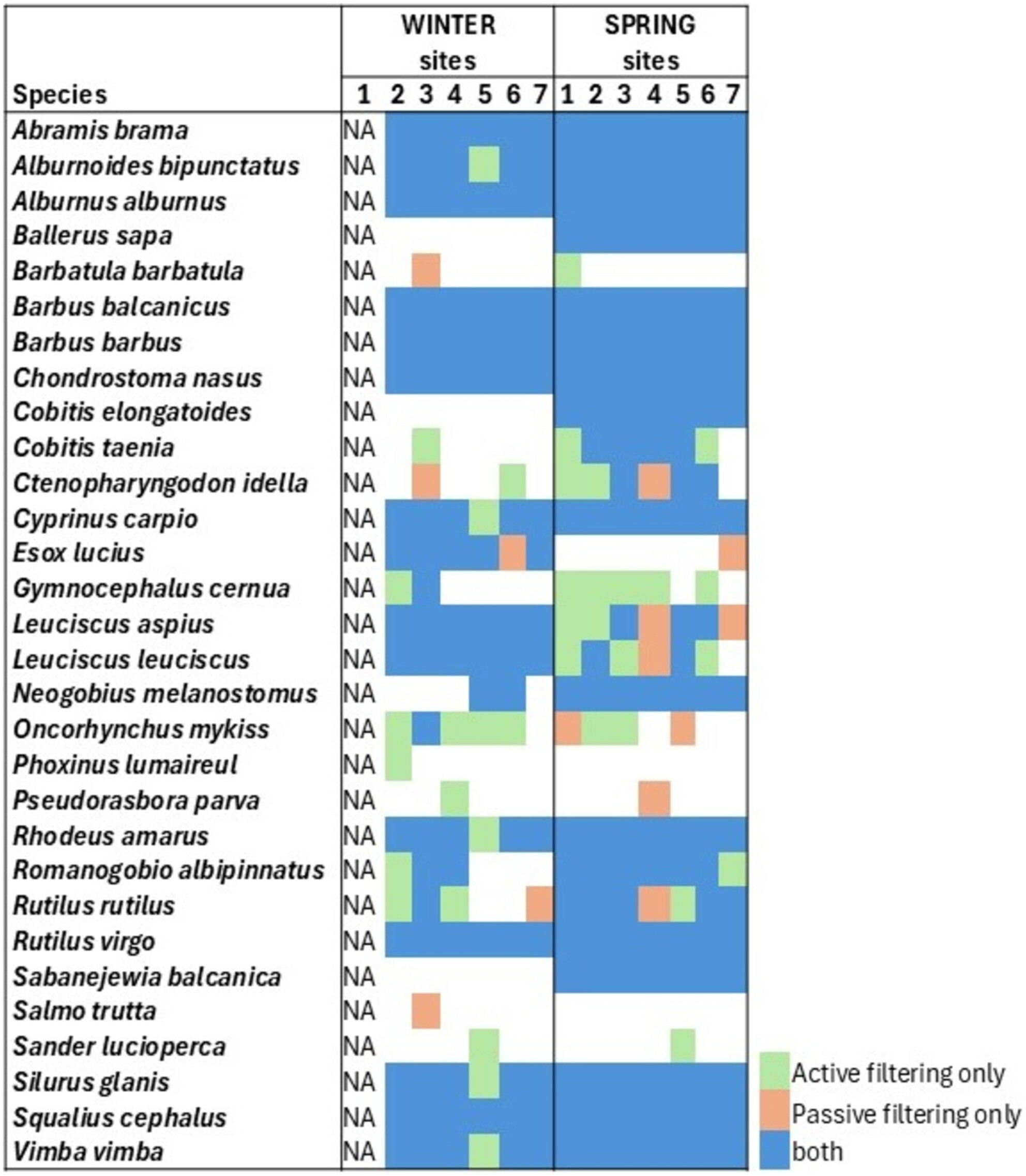
Passive eDNA collection using coffee filters is a robust and scalable method for biodiversity monitoring in both large rivers and small streams. Both eDNA sampling methods, active eDNA filtering, and passive eDNA collection, detected a greater fish species richness per site than electrofishing. Significant differences in species read abundance were observed between upstream and downstream sites, with a marked increase during spawning periods.
Monitoring of the Invasive Round Goby (Neogobius melanostomus) in an Estuarine Seascape Based on eDNA
- First Published: 08 March 2025
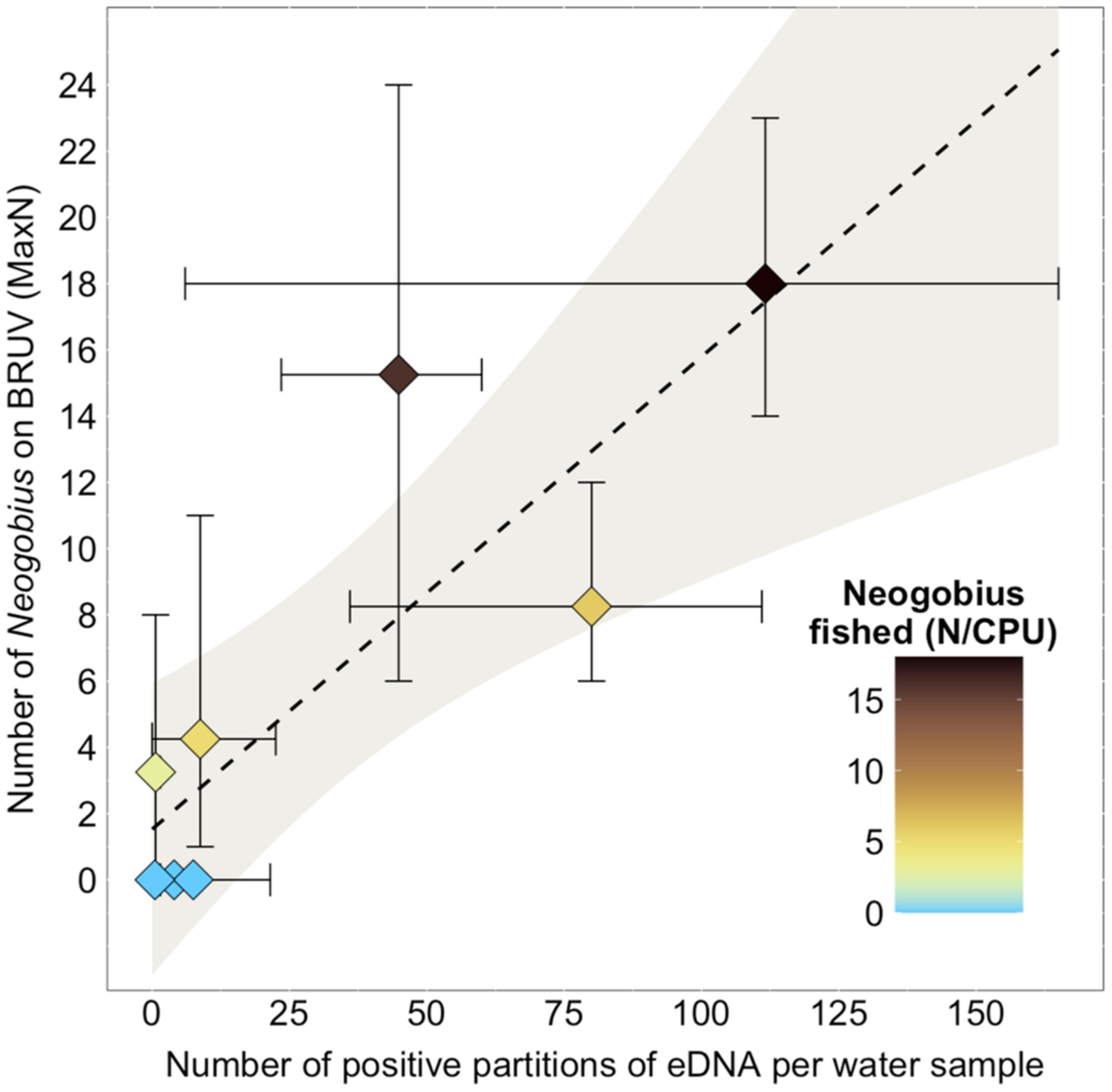
Effective monitoring of marine invasive species is crucial for early detection valuable to management. In this study, we develop and evaluate environmental DNA monitoring to assess the presence of the round goby fish (Neogobius melanostomus) in a seascape environment. We found that with a half-life of 9.8 h in 15 PSU salinity and 15°C temperature and eDNA presence of the species confirmed with fishing and video, it is possible to monitor the current presence and abundance to support conservation management.
Environmental Matrices Need Consideration: Insights From Water and Biofilm Environmental DNA for Multi-Taxonomic Biomonitoring
- First Published: 20 March 2025
An Optimized Probe-Based qPCR Assay for the Detection and Monitoring of the Invasive Lionfish (Pterois volitans) in the Atlantic
- First Published: 14 March 2025
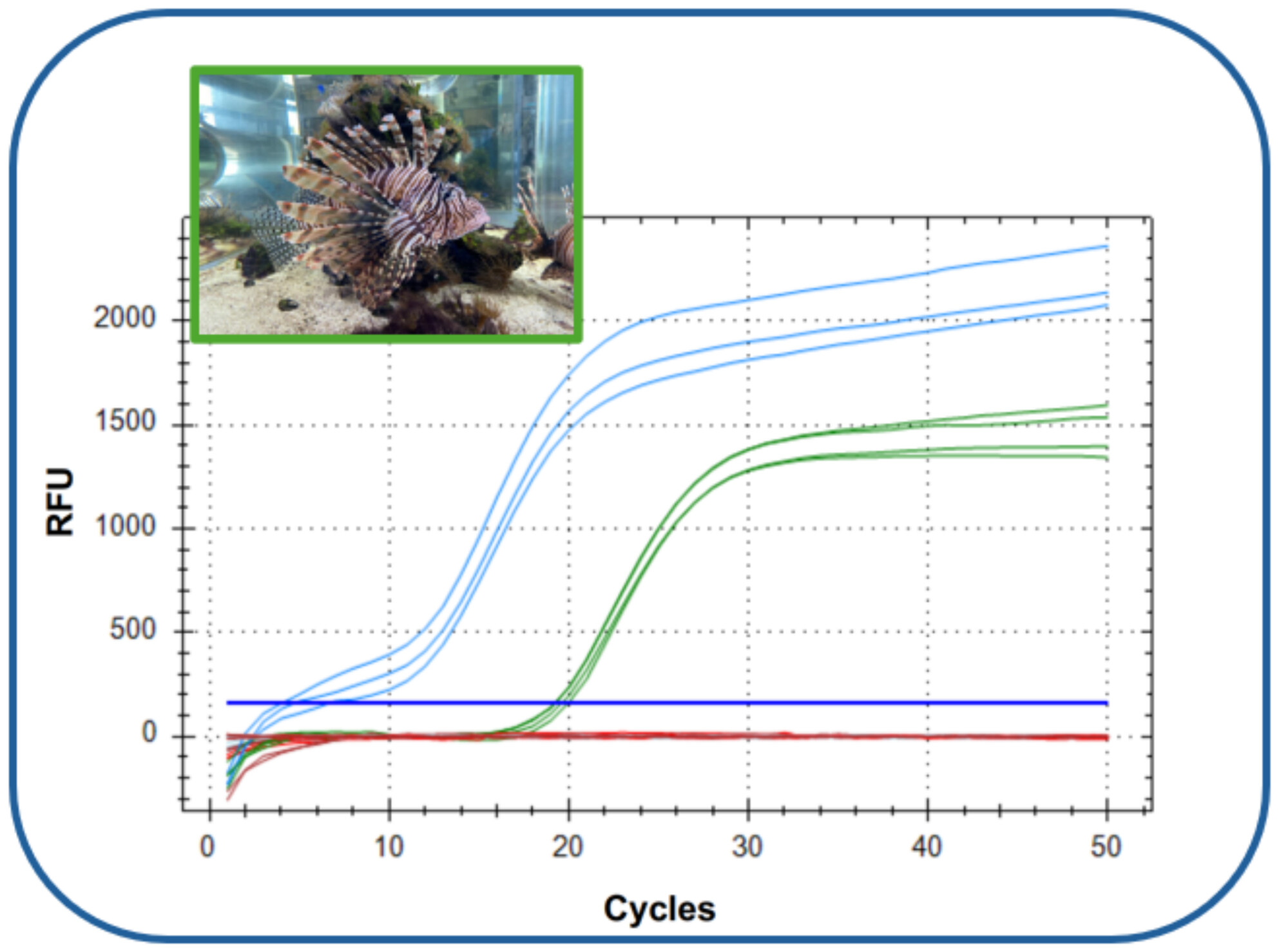
Lionfish are invasive in the western Atlantic, with populations expanding rapidly since their introduction in the 1980s. Here we evaluate the specificity of the currently available lionfish traditional PCR assay and provide an alternative, more sensitive qPCR assay for the detection of lionfish in the western Atlantic region.
Environmental RNA-Based Surveillance of Viral Hemorrhagic Septicemia Virus (VHSV) in Olive Flounder (Paralichthys olivaceus) Aquaculture: Detection Dynamics and Risk Assessment
- First Published: 20 March 2025
Metazoan Diversity and Its Drivers: An eDNA Survey in the Pacific Gateway of a Changing Arctic Ocean
- First Published: 24 March 2025
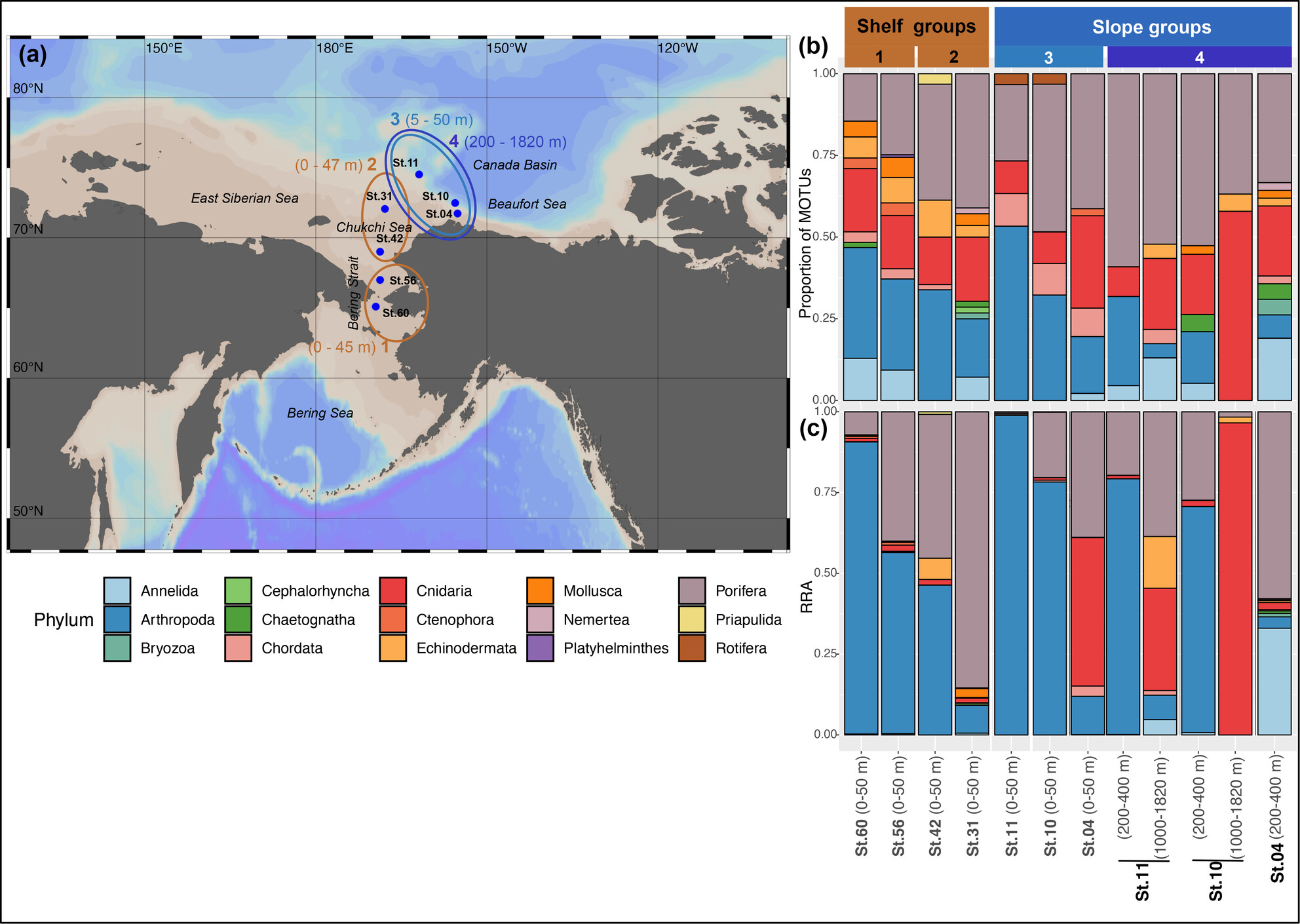
In the face of climate change, organisms must adapt or shift their ranges to survive. Our study conducted the first COI marker-based eDNA survey in the Pacific Gateway of the Arctic Ocean, analyzing seawater samples from the Bering Strait and Chukchi Sea. We detected diverse metazoan taxa, identified indicator species, and linked community composition to environmental variables, noting that warmer temperatures might enhance alpha diversity due to the influx of warmer, fresher water, while colder, saltier water-associated taxa may be negatively impacted by ongoing environmental change.
PERSPECTIVE
A Framework to Unify the Relationship Between Numerical Abundance, Biomass, and Environmental DNA
- First Published: 02 March 2025
ORIGINAL ARTICLE
Squishy and Crunchy Invasive Invertebrates: Environmental DNA Is Not Shed Equally
- First Published: 20 March 2025
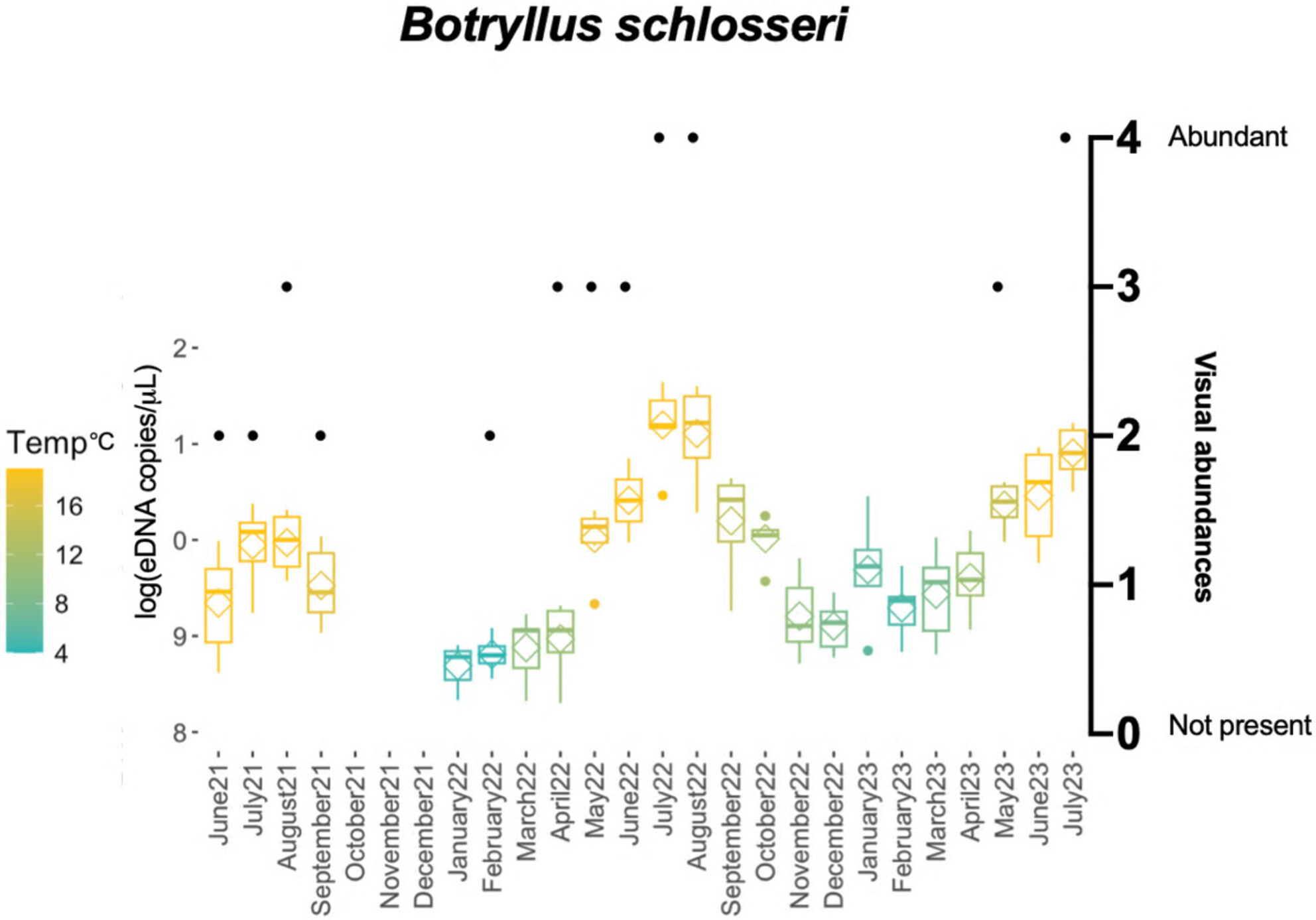
Environmental DNA has been used to detect invasive species in aquatic ecosystems with varying degrees of success. Here, using laboratory experiments and a two-year time series, we found that squishy species (those lacking an exoskeleton) generally shed eDNA consistently with their abundance, whereas crunchy species (those with a shell or exoskeleton) do not. Thus, eDNA signals cannot be assessed equally for all taxa, especially invertebrates with varied bauplans.
Persistent Gaps and Errors in Reference Databases Impede Ecologically Meaningful Taxonomy Assignments in 18S rRNA Studies: A Case Study of Terrestrial and Marine Nematodes
- First Published: 25 March 2025
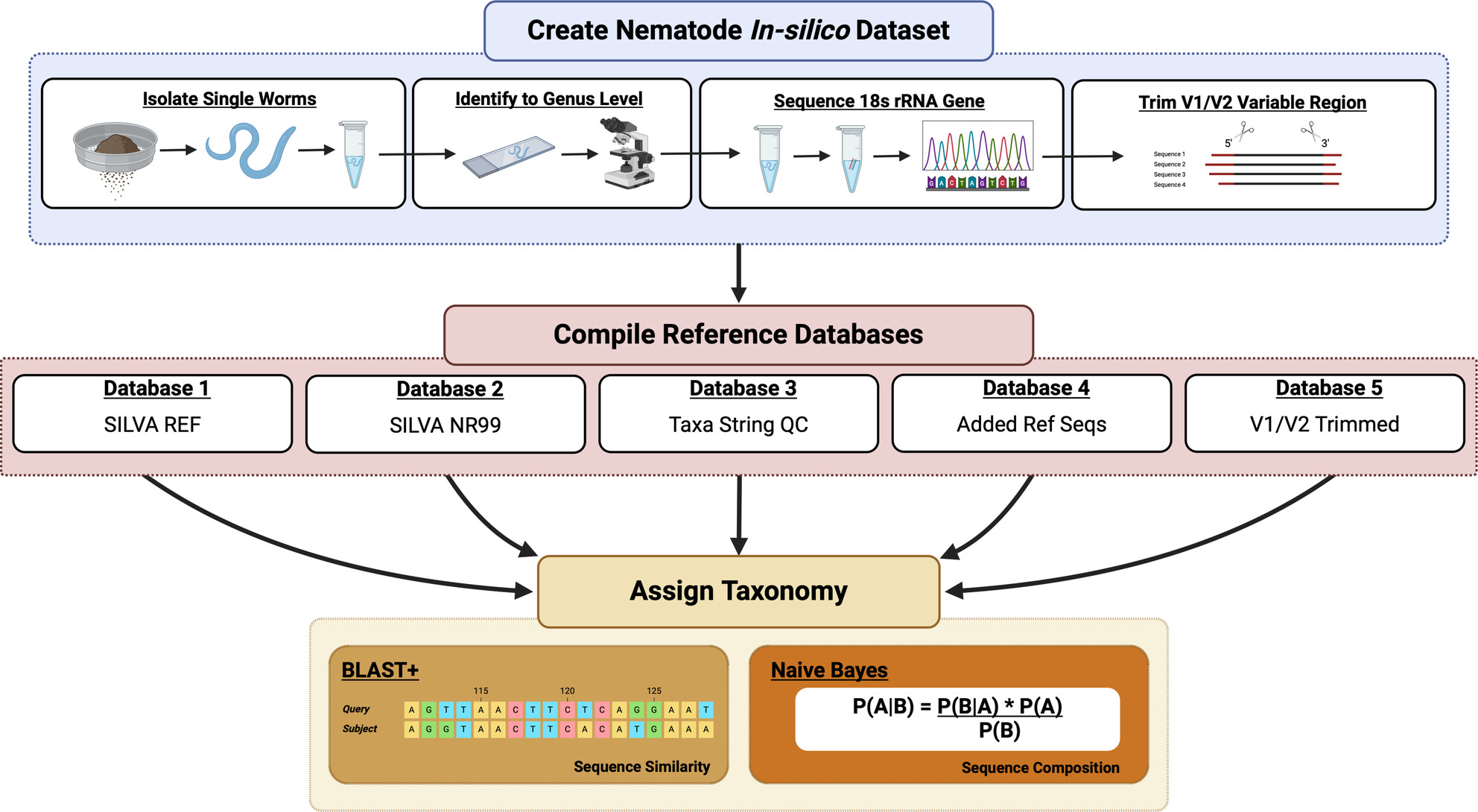
Public molecular databases (i.e., SILVA, EUKARYOME, BOLD) for most microbial metazoan phyla (nematodes, tardigrades, kinorhynchs, etc.) are sparsely populated, negatively impacting our ability to assign ecologically meaningful taxonomy to these understudied groups. We use two in silico datasets to show that metabarcoding taxonomy assignments using the 18S rRNA gene can be dramatically improved by maximizing phylogenetic sampling in public reference databases and curating Linnaean taxonomy strings associated with each reference sequence. Our results highlight the urgent need for phylogenetically informed expansions of public reference databases, focused on poorly sampled lineages which are now robustly recovered via eDNA metabarcoding approaches.
Reconstruction of Marimo Population Dynamics Over 200 Years Using Molecular Markers and Fossil Plankton Remains
- First Published: 31 March 2025
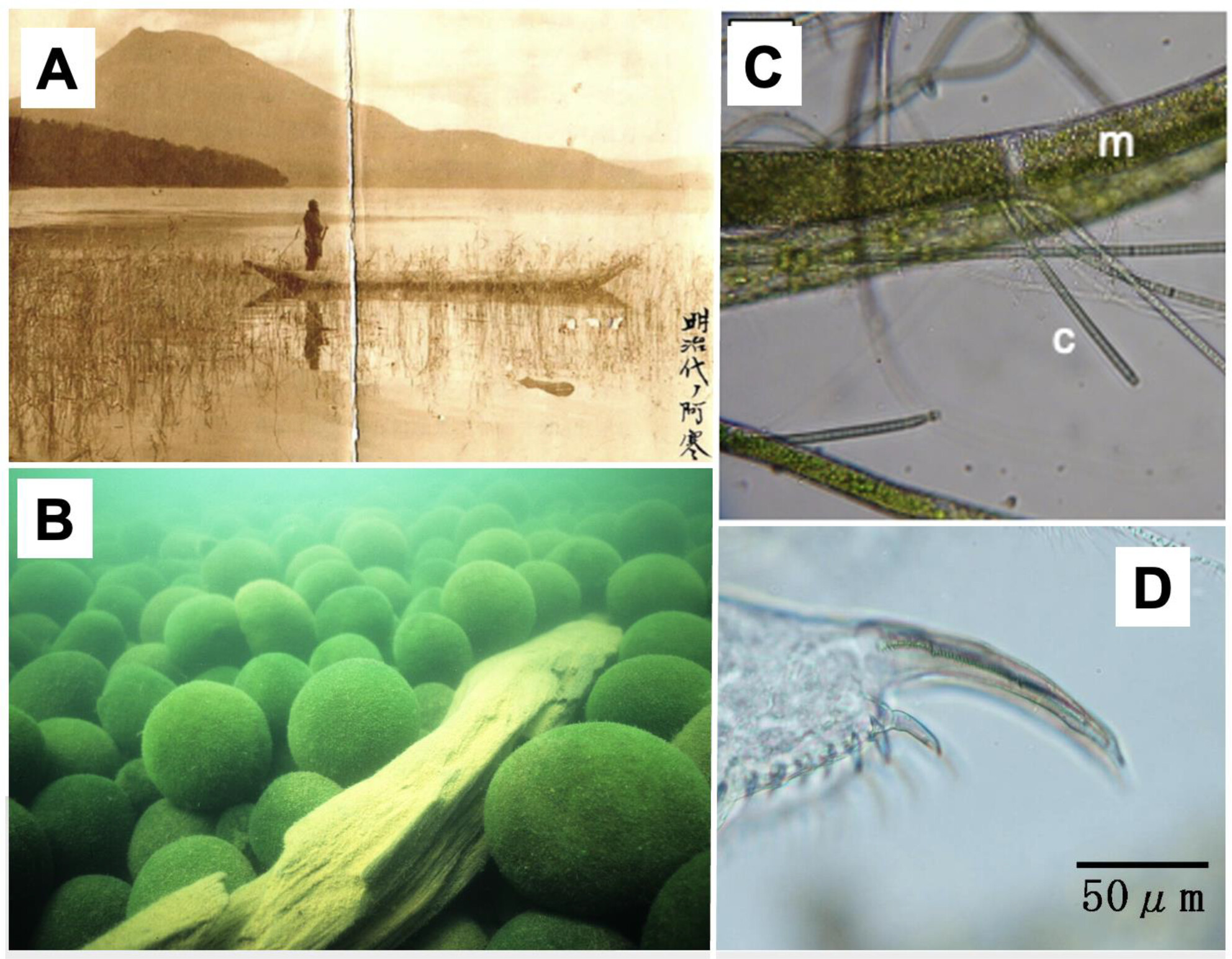
We introduce a new method utilizing environmental DNA to accurately reconstruct past population abundances of organisms that do not fossilize, and applies this method to estimate the abundance of the large marimo, spherical colonies of the green alga Aegagropila linnaei in Lake Akan. Because marimo are iconic organisms in freshwater ecosystems, this study will resonate with scientists in not only ecology in general but also those interested in the use of sedDNA in paleolimnology, especially.
The First eDNA-Based Assessment at the World's Most Remote Inhabited Islands: Investigating Marine Vertebrate Diversity at Tristan da Cunha
- First Published: 26 March 2025
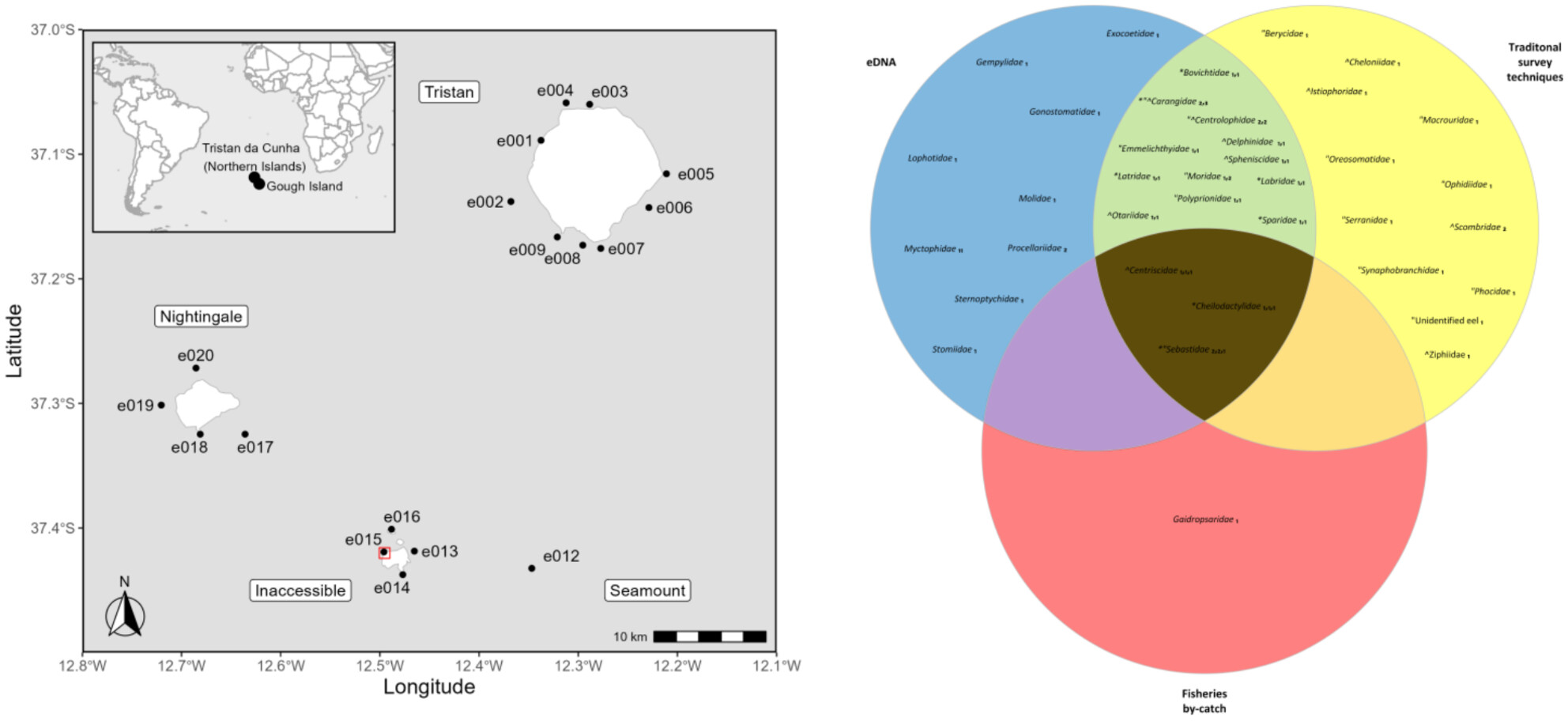
This study employed eDNA to characterize marine vertebrate biodiversity at Tristan da Cunha, the world's most remote archipelago. The multi-assay approach successfully detected 51 Operational Taxonomic Units (OTUs) encompassing 24 families, 32 genera, and 13 species, resolving the islands characteristic diversity profile, and also identifying rare and vulnerable taxa underrepresented by traditional surveys, and identifying species previously unrecorded at the islands. eDNA resolved greater species richness in kelp compared to non-kelp habitats.
METHOD
Challenges in Detecting Ecological Interactions Using Sedimentary Ancient DNA Data
- First Published: 01 April 2025
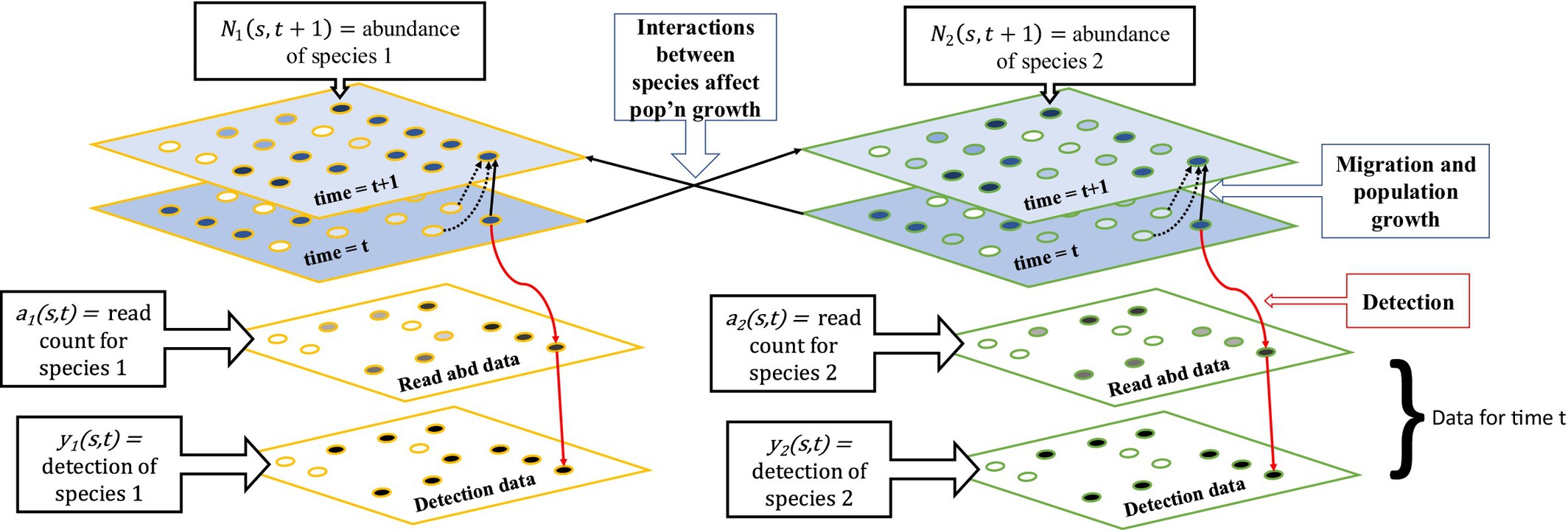
Sedimentary ancient DNA data are often used to infer associations between species, which can generate hypotheses about biotic interactions, a key part of ecosystem function and biodiversity science. We have developed a realistic simulation to evaluate five common methods from different fields for this type of inference. We find that across all methods tested, false discovery rates of interspecies associations are high under realistic simulation conditions, indicating a need for further testing and method development.
ORIGINAL ARTICLE
Breaking the Standard: Can Oxford Nanopore Technologies Sequencing Compete With Illumina in Protistan Amplicon Studies?
- First Published: 28 March 2025
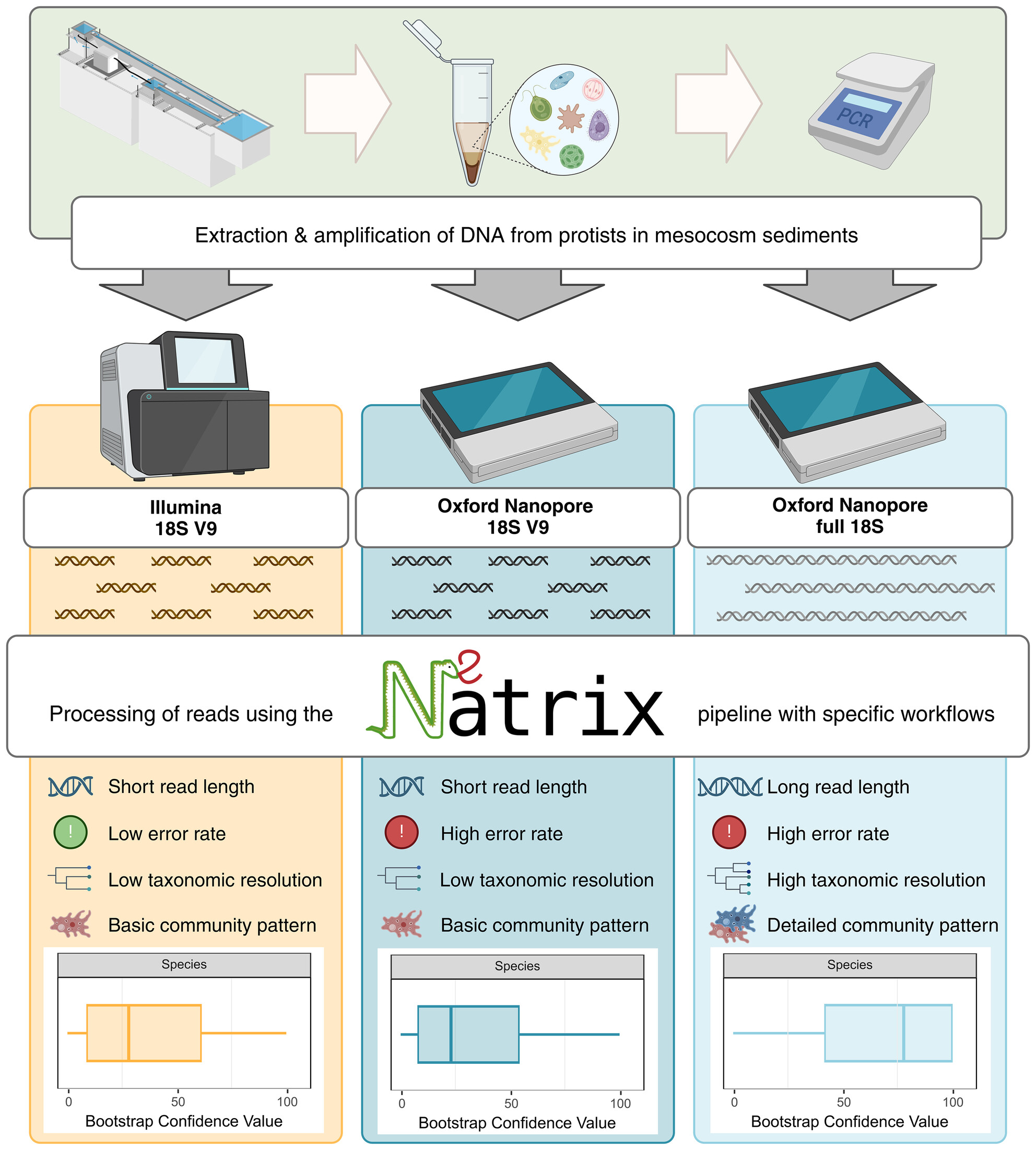
Full-length 18S Oxford Nanopore sequencing improves taxonomic resolution and reveals subtle species responses in sediment protists, outperforming Illumina V9 sequencing despite higher error rates. (Created in BioRender. Bludau, D. (2025) https://BioRender.com/0ura1xc).
Water Volume, Biological and PCR Replicates Influence the Characterization of Deep-Sea Pelagic Fish Communities
- First Published: 28 March 2025
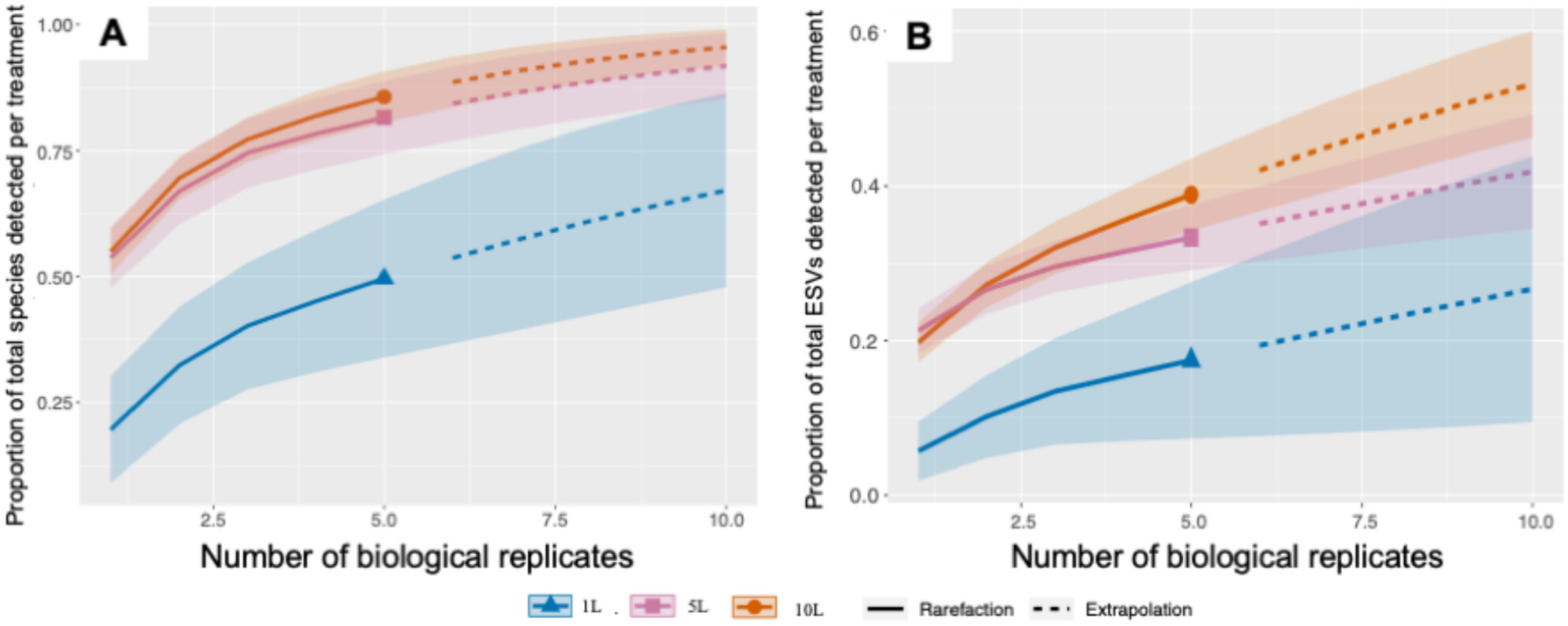
We investigated the effects of sampled water volume, biological, and PCR replicates in characterizing deep-sea pelagic biodiversity. We suggest that future studies collect at least 5 L, 5 or more field replicates, and 5–10 PCR replicates to adequately investigate deep-sea pelagic biodiversity when using Niskin bottles coupled with CTD. Our study provides guidance for future eDNA studies and a potential route to expand eDNA studies at a global scale.
Monitoring Atlantic Salmon (Salmo salar) Smolt Migration in a Large River System Using Environmental DNA
- First Published: 01 April 2025
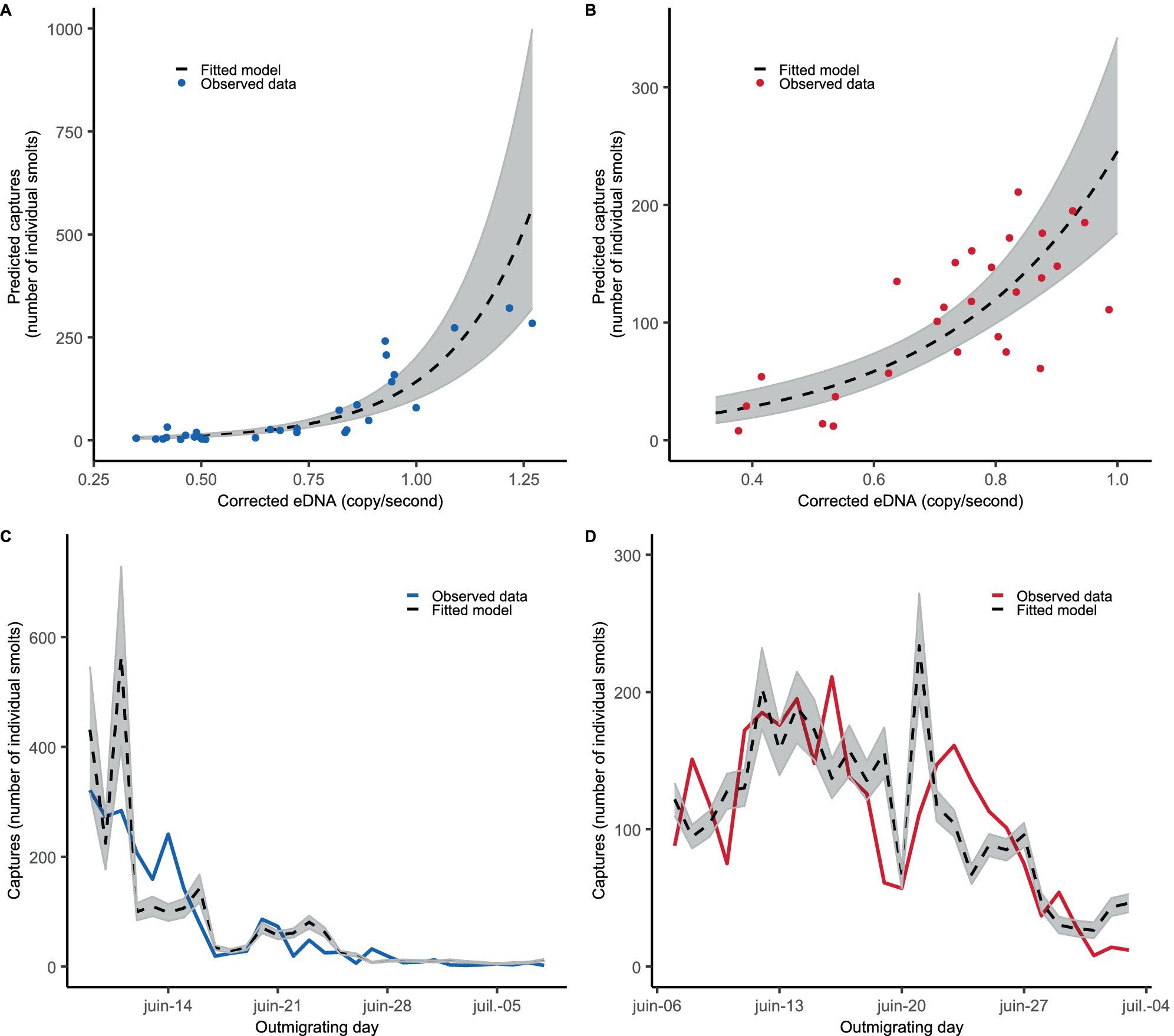
In the context of an important Atlantic salmon restoration program conducted in a large river system, we took advantage of the yearly count of smolt to evaluate the suitability of eDNA as a semi-quantitative tool to monitor smolt migration. Results show a positive and exponential relationship between eDNA concentration and the number of caught smolts each day.
SPECIAL ISSUE ARTICLE
Combining eDNA Metabarcoding, Hydrology-Based Modeling and Camera Trap Datasets to Assess the Potential of River eDNA in Monitoring Terrestrial Mammals
- First Published: 05 April 2025
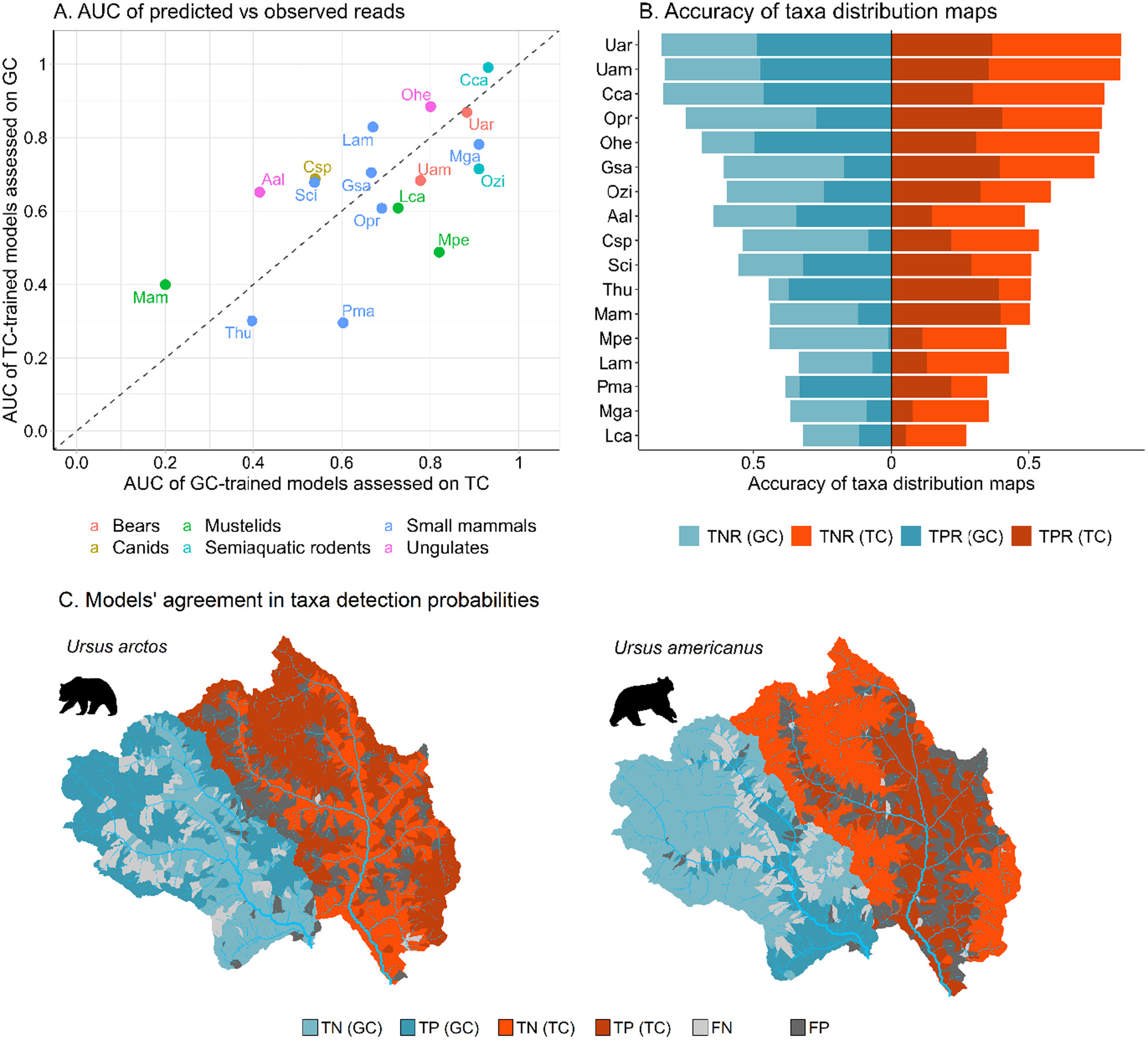
By applying the hydrology-based eDITH model to two mountainous catchments and comparing it with camera trap data, we found that for 9 out of 15 taxa, predicted distributions predominantly matched camera trap observations. We utilized predicted eDNA patterns to compare the potential of different sampling strategies and found that monitoring biodiversity at the landscape scale can be achieved by predominantly sampling downstream reaches. This study highlights the potential of river eDNA and hydrology-based models to enhance the monitoring of terrestrial mammals.
ORIGINAL ARTICLE
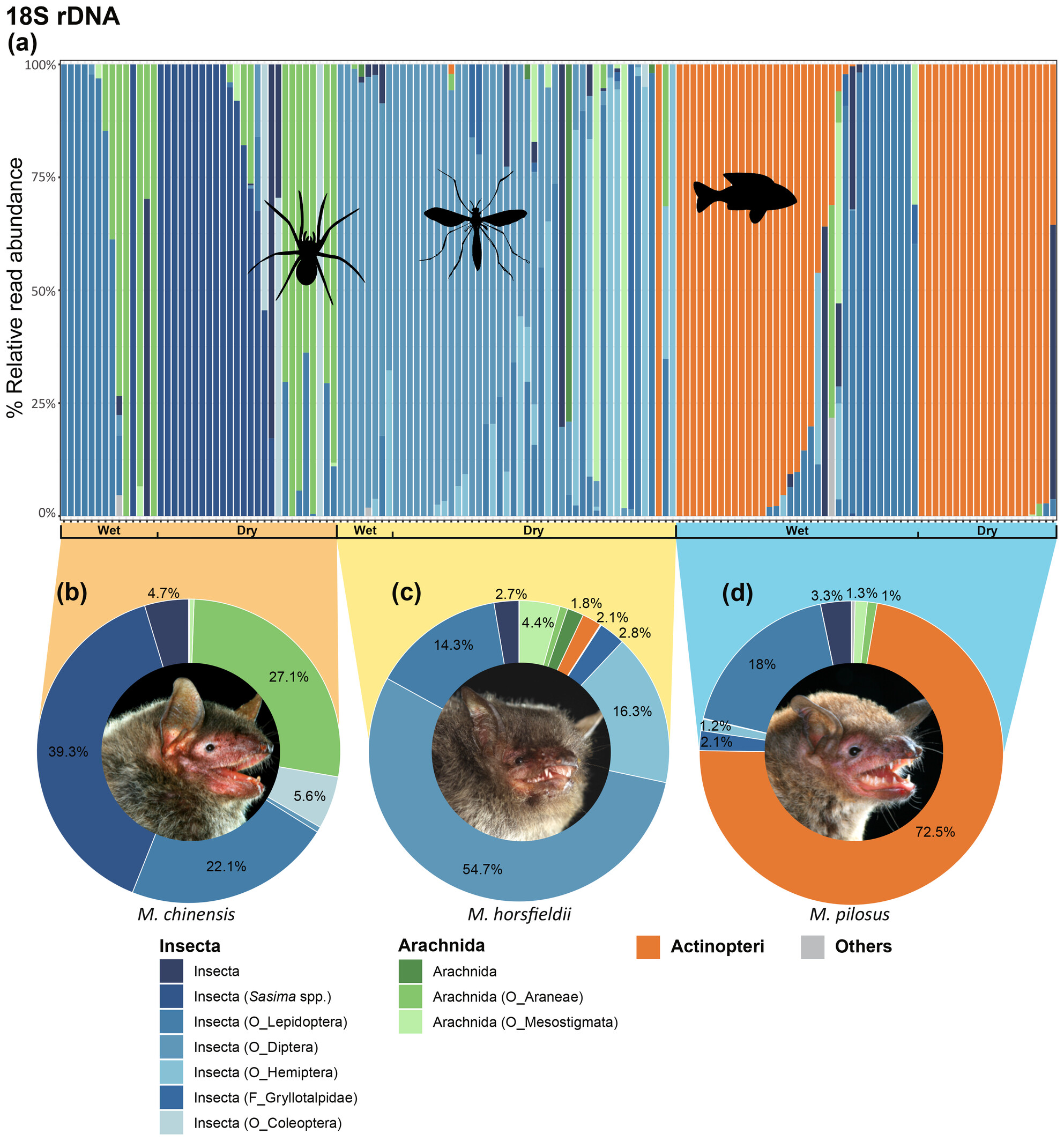
Foraging Niche Partitioning of Three Myotis Bat Species and Marine Fish Consumption by Myotis pilosus in a Subtropical East Asian Region
- First Published: 05 April 2025
In the context of an important Atlantic salmon restoration program conducted in a large river system, we took advantage of the yearly count of smolt to evaluate the suitability of eDNA as a semi-quantitative tool to monitor smolt migration. Results show a positive and exponential relationship between eDNA concentration and the number of caught smolts each day.
Development, Validation, and Implementation of eDNA-Focused qPCR Assays to Detect and Distinguish Between Goldfish (Carassius auratus) and Prussian Carp (Carassius gibelio)
- First Published: 05 April 2025
Of Islands on Islands: Natural Habitat Fragmentation Drives Microallopatric Differentiation in the Context of Distinct Biological Assemblages
- First Published: 08 April 2025
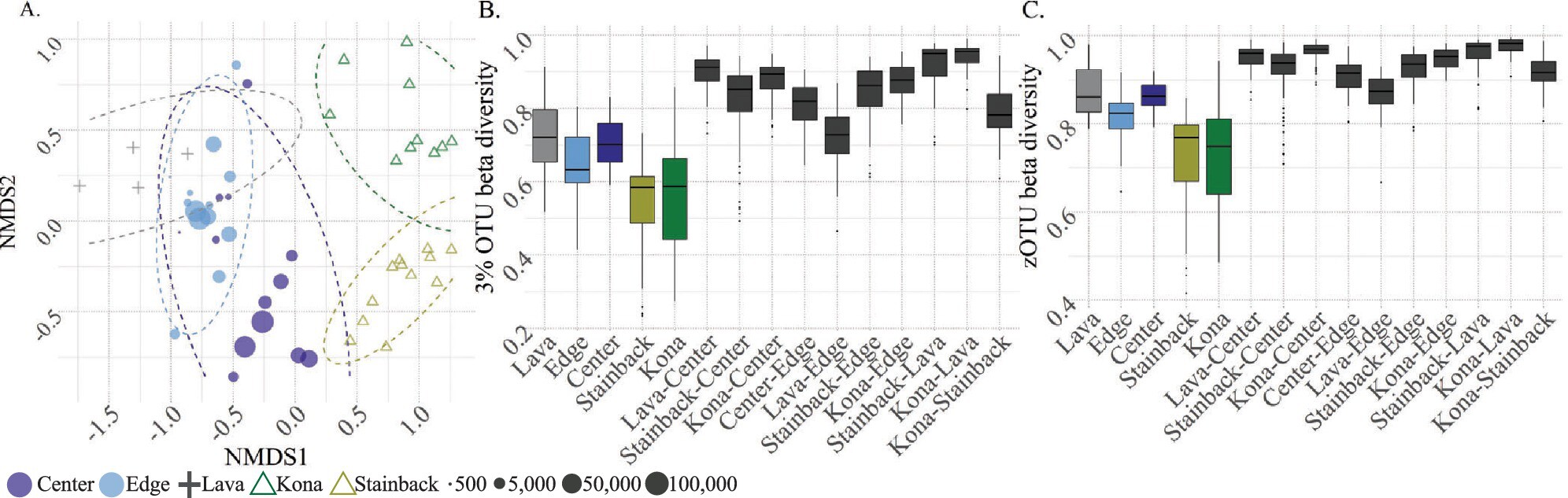
Leveraging a natural experiment provided by arthopod communities in Hawaiian wet forest fragments isolated by lava flows, this study examines the impact of natural habitat fragmentation on species-equivalent (3% radius OTUs) and haplotype-equivalent (zOTUs) scales. The authors observe that fragments host distinct communities from native forest and experience higher rates of turnover, both at species- and haplotype-equivalent scales. This system provides an exciting opportunity to test the idea of natural habitat fragments as “crucibles of evolution,” given that we find arthropods to be undergoing genetic differentiation while isolated in different biological communities.
Linking Water to the Bottom: eDNA Study of Benthic Invertebrates and Invasive Species in the Venice Lagoon
- First Published: 09 April 2025
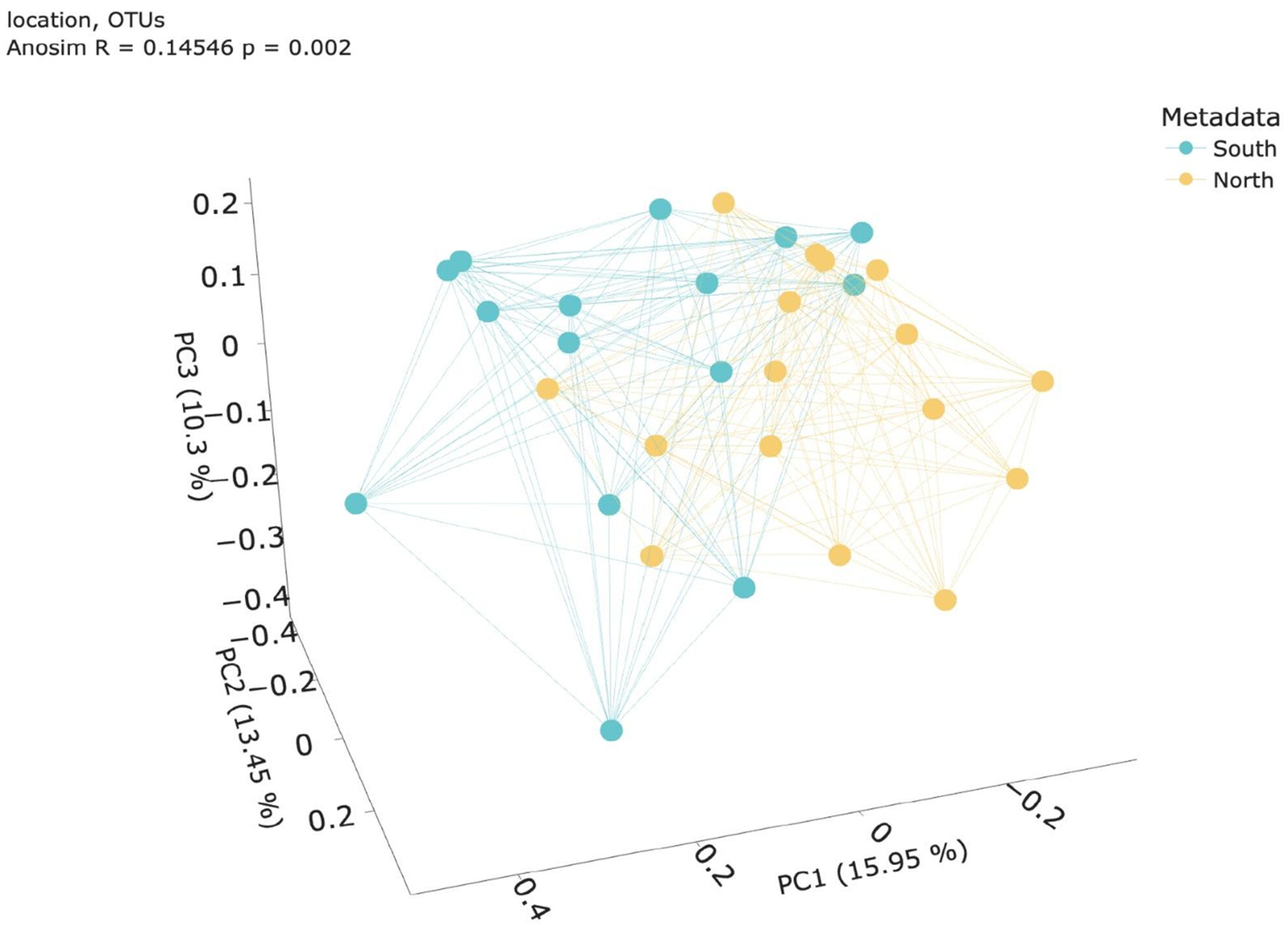
This study used environmental DNA (eDNA) metabarcoding of superficial water samples to assess benthic invertebrate biodiversity in the Venice Lagoon. Sampling from surface water revealed 80 taxonomic units, mostly benthic species, including several invasive ones, with significant spatial and seasonal variations in community composition. The results demonstrate that eDNA from surface water can effectively monitor benthic biodiversity, providing a valuable tool for ongoing assessments in lagoons and similar environments.
Comparative Analysis of Environmental DNA Metabarcoding and Spectro-Fluorescence for Phytoplankton Community Assessments
- First Published: 16 April 2025
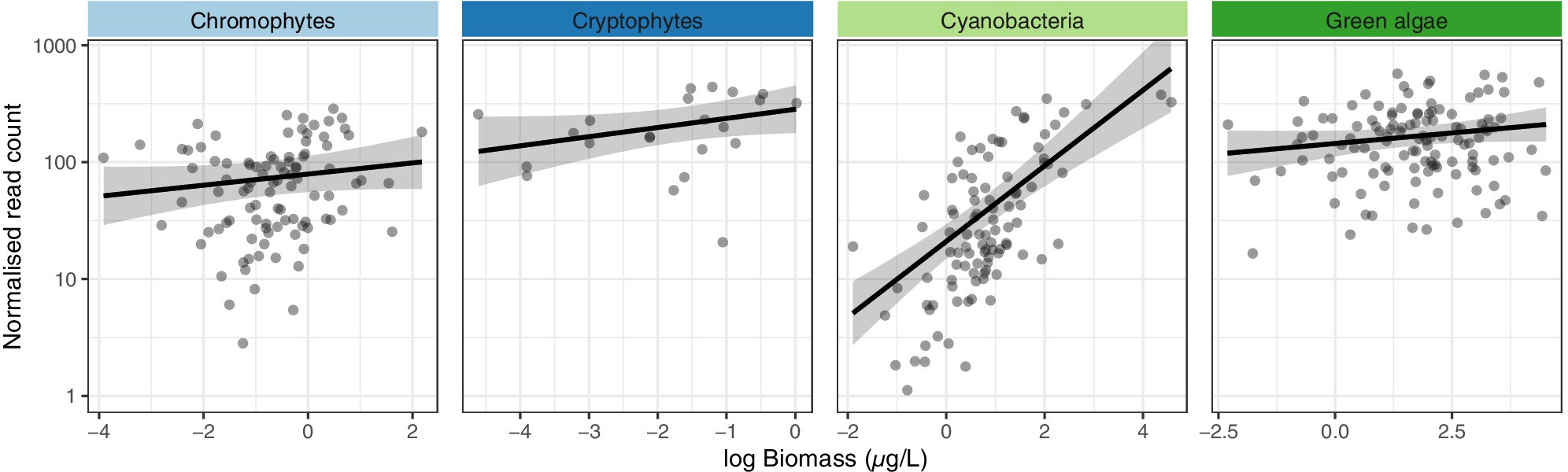
We conducted a comparative analysis of phytoplankton community composition using fluorescence-based measurements and DNA metabarcoding in a controlled mesocosm experiment. A strong correlation was observed between fluorescence-derived biomass and DNA read abundance for cyanobacteria, but not for other phytoplankton groups. While DNA metabarcoding may not provide reliable biomass estimates for the majority of phytoplankton groups, it was systematically better at detecting cryptophytes, offers higher taxonomic resolution, and the capability to detect rare phytoplankton.
Assessing the Relationship Between Sedimentary Microbial Community Structure and Pollutants in Stormwater Wetlands Using eDNA Metabarcoding
- First Published: 17 April 2025
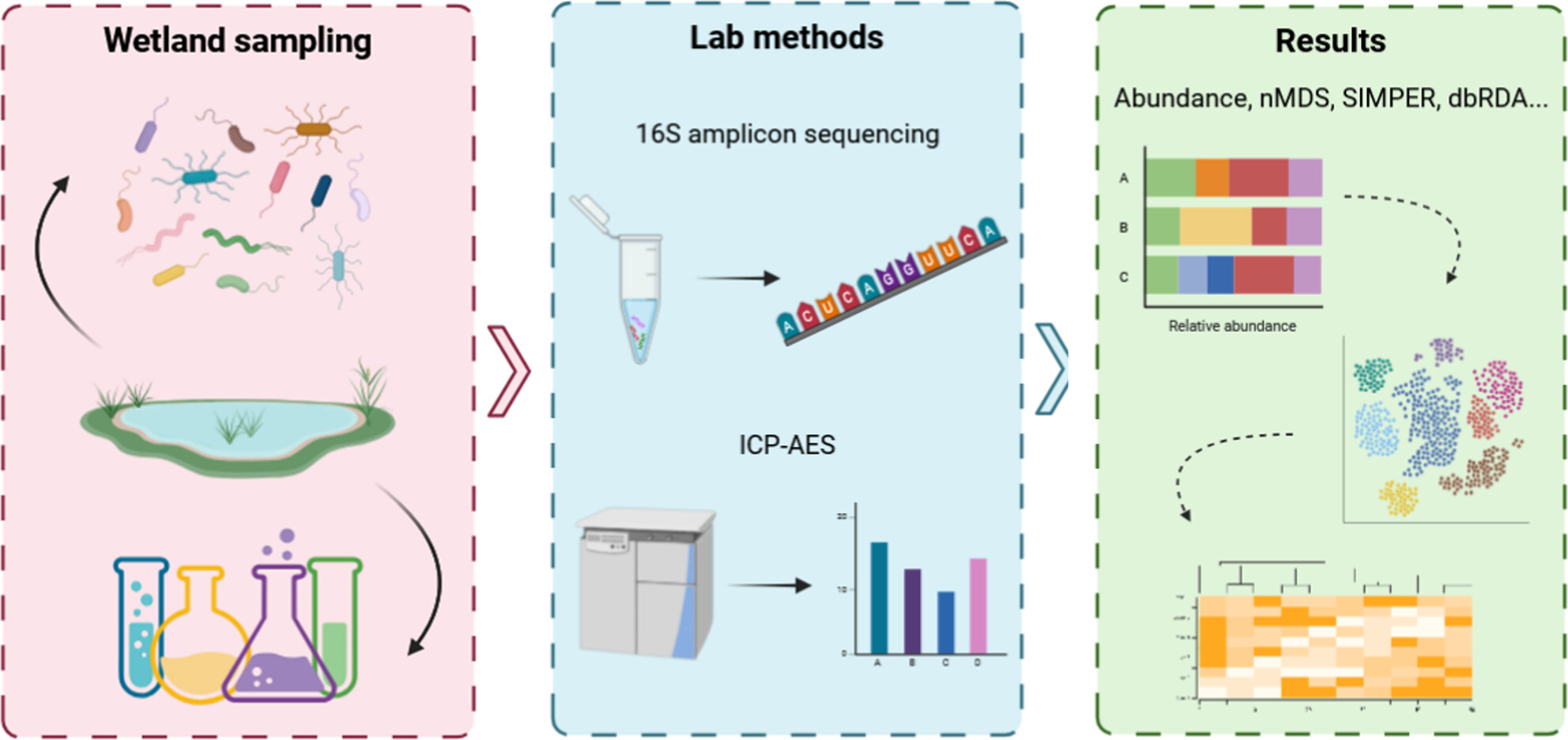
Constructed wetlands are designed to retain contaminants from stormwater which would otherwise pose significant threats to freshwater ecosystems downstream. Here, we utilized the ability of environmental DNA metabarcoding to assess biological profiles quickly and reliably to characterize the sedimentary microbial profiles of nine urban stormwater wetlands and one rural wetland, and compare against their chemical profiles. We found significant variation between the microbial communities driven primarily by Cyanobacteria and Proteobacteria, and identified zinc and barium as key chemicals influencing microbial community structure.
Metabarcoding Versus Species-Specific Primers to Estimate Salmo trutta Biomass and Density in Mountain Streams
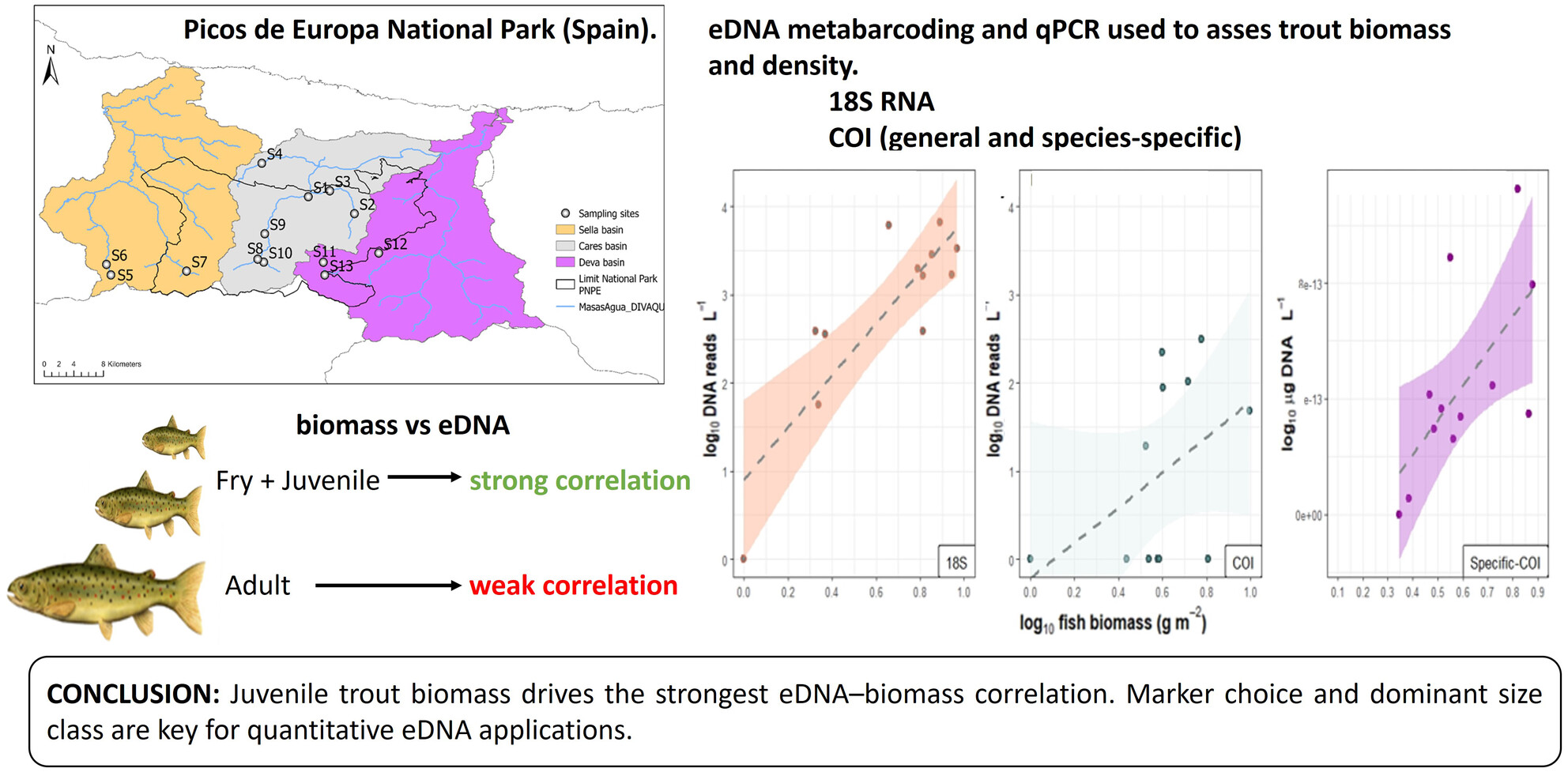
- First Published: 21 April 2025
This study explores the use of environmental DNA (eDNA) as a noninvasive, cost-efficient tool to monitor trout populations in mountain streams. It found that eDNA concentration correlates positively with trout biomass and density, particularly using general primers for 18S rRNA gene and COI-Salmo trutta specific primers. The biomass or densities in each size class (fry, juvenile, and adult) of brown trout contribute differently to DNA quantity in stream water. Juvenile trout biomass contributed most significantly to the observed eDNA-biomass relationship, suggesting eDNA is a promising method for fish population management and conservation.














