Environmental DNA (eDNA) Quantitative Polymerase Chain Reaction-Based Assays for Surveying 125 Taxa of Importance to North America
Funding: This study was supported by Genome Canada's Large-Scale Applied Research Project (LSARP) program #312ITD, which also includes funding from Genome Québec and Genome British Columbia to V.S.L. and C.C.H. and the Canada's Research Chair Program to V.S.L. Additional support was from a NSERC collaborative research and development grant #CRDPJ 531846-18 to C.C.H., M.J.A. and N.A.P. are recipients of NSERC CGS-D graduate fellowships. M.L.D.L. is a Liber Ero Fellow.
ABSTRACT
Timely and accurate assessment of the presence of at-risk or invasive species is critical for effective responses to climate change and human impacts. For example, at-risk species are often difficult to find, while invasive species are often well established before their infiltration is detected using conventional surveying methods. However, all organisms release genetic material such as DNA into their surroundings, leaving traces of themselves that can be detected using environmental DNA (eDNA) methods. These approaches are powerful tools in the conservation toolbox, as they are transforming how risk assessments and the evaluation of mitigation and remediation effectiveness are done. Despite this, poorly performing tools hinder broad adoption of eDNA-based detection methods, due in part to their associated high false negatives and false positives that can impair effective management decision-making. iTrackDNA is a multi-year, large-scale applied research project that is addressing these concerns with researchers and end users from various sectors across North America. It is building end-user capacity through innovative, accessible, socially responsible genomics-based analytical eDNA tools for effective decision-making by publishing 125 quantitative real-time polymerase chain reaction (qPCR) primer/probe sets designed to detect key invertebrates, fish, amphibians, birds, reptiles, and mammals in coastal and inland ecosystems important to North America, with an emphasis on Canada. These 125 assays were designed to meet or exceed the new Canadian Standards Association (CSA) consensus-based and multi-stakeholder national standards for eDNA (CSA W214:21 and CSA W219:23). Herein, we describe how we applied eDNA assay design and validation approaches across a wide range of animal taxa to achieve compliance.
1 Introduction
Natural resource decisions require reliable information on animal taxa to assess, monitor, and mitigate the deleterious impacts on biodiversity and water quality (Ulicsni et al. 2019). The use of environmental DNA (eDNA), that is, the genetic material released by organisms into their surroundings, for environmental monitoring and risk assessments can provide non-destructive, cost-effective, and accurate information on species biodiversity. Moreover, certain conventional methods are being replaced by eDNA standardized protocols. For example, this has already been established in some mandatory protocols to inventory key species requested by governments (e.g., great crested newt (Buxton et al. 2022), turtles (Gouvernement du Québec 2022), endangered fish (Gouvernement du Québec 2023), at-risk snakes (Gouvernement Du Québec 2024a), and some salamander species (Gouvernement Du Québec 2024b, 2024c)). These eDNA-based protocols will continue to gain popularity as they are robust and cost-effective for conservation biology.
Advances in molecular technologies have greatly improved our ability to survey global biodiversity in natural ecosystems and impacted areas. Rising societal pressure to balance environmental protection with natural resource extraction and increasingly stringent regulatory requirements necessitate that countries support various stakeholders (i.e., regulators, industry, Indigenous Peoples, and non-governmental organizations (NGOs)) with new tools to monitor the biosphere, build overlapping datasets to monitor entire ecosystems, and decrease time to action (Fitzgerald et al. 2021). In addition, ecological effects assessment and trend monitoring systems need overhauling to increase effectiveness, resolution, and efficiency. Reliable, socially responsible, and timely monitoring techniques of targeted taxa population status in coastal and inland ecosystems are not readily accessible (Van Der Heyde et al. 2020).
Several large international-based research projects, such as GEN-FISH (https://gen-fish.ca/fr/), BIOSCAN (https://ibol.org/bioscan/), and iTrackDNA (https://itrackdna.ca/) are contributing to eDNA methods development to improve biodiversity survey results with reduced invasiveness. GEN-FISH is working on toolkits for the identification of freshwater fish species and the expression of stress-related genes in parts of Canada and the USA. For over a decade, BIOSCAN has built DNA barcode reference libraries to assess global biodiversity. iTrackDNA provides international leadership in genomics and the social sciences to ensure rigor and reliability with transformative, socially responsible eDNA methods. iTrackDNA also builds capacity by bringing innovative tools to support the standardization of environmental data that can be fed into industry operational and risk assessment models to de-risk decisions while decreasing time to action. Indeed, all these large-scale genomic projects aim to overcome the current barriers to eDNA uptake by end-users, such as inconsistent practices, poor-quality eDNA detection tools, and unacceptably high false negatives and false positives that can compromise effective management decision-making (Ruppert et al. 2019; Langlois et al. 2021).
The most widely used method for detecting eDNA of targeted species or taxonomic groups is quantitative real-time polymerase chain reaction (qPCR). qPCR equipment is widely available and relatively accessible in comparison with other eDNA methods. While targeted species assay results are the most directly translatable for regulatory purposes, genus- or family-level assays can act as general screens for rapid, cost-effective analyses in scenarios such as determining if fish inhabit an impacted area, if mouse-eared bats inhabit abandoned mines, or if a parasite host (deer for brainworm) or invasive mussels are present (all these examples have been covered by eFish1, eMyotis1, eOdocoileus2, and eDrspp7 assays, respectively, described below).
However, there is a wide range in the quality of qPCR-based assays within the published literature and in practice (Taylor et al. 2019). That is why most research projects are attempting to address important quality assurance/quality control (QA/QC) aspects that are required for confident uptake of targeted eDNA detection assays, namely standardization and assay validation within the context of qPCR kits. The iTrackDNA project received feedback from 50+ project participants representing provincial and federal regulators, industry, First Nations, and NGOs on priority species for which eDNA assays were desired. As a result of this consultation, a list of over 100 species was generated. From this priority list, 125 qPCR-based eDNA assays were designed and validated using a workflow described previously (Langlois et al. 2021). These assays cover the detection of 59 distinct fish, 25 mammals, 21 herptiles (amphibians and reptiles), 18 invertebrates, and two bird taxa, including at-risk, indicator, invasive, and socio-economically important organisms.
The eDNA assays reported herein were designed to meet two Canadian national eDNA standards. These two documents, published by the Canadian Standards Association (CSA), present consensus-based, multi-stakeholder national standards to define minimum criteria that need to be met to have confidence in eDNA data (Gagné et al. 2021; Abbott et al. 2023). The first standard applies to the reporting of information regarding the conducting of eDNA biological surveys or assessments, while the second defines the performance criteria of targeted qPCR assays used in eDNA surveys. According to the CSA standard, those qPCR assays must be comprised of primers and a probe rather than primers combined with an intercalating agent, as the latter assay configuration does not have the desired level of performance (Abbott et al. 2023).
The degree of inclusion of eDNA in regulatory protocols depends on the availability of appropriate and quality tools, standards, and regulatory approvals (Langlois et al. 2021). This critical need to develop standards-compliant eDNA assays will help fill knowledge gaps in eDNA research so that the use of eDNA can be included in regulatory decisions to improve biodiversity monitoring and conservation management efforts worldwide. The aim of the present paper is to provide an example of the approach taken by the iTrackDNA project to generate 125 robust eDNA assays that are compliant with the CSA standards and generally meet the highest validation level as previously described (Langlois et al. 2021; Thalinger et al. 2021). While we continually strive to ensure robust assays and validation activities are ongoing, it is important to note that some assays herein are presented with limited validation due to constraints in mitochondrial sequence information, access to voucher specimens, and/or field samples. For example, we were not yet able to establish that the yellow perch (Perca flavescens) ePEFL1 assay does not detect other Percidae species that could be confounders in vitro due to lack of access to appropriate voucher specimens (File S2). As with any eDNA assay, it is imperative that users ensure that appropriate checks are incorporated within the specific contexts in which the assay is to be used and that appropriate detection of target species within the survey area is established.
2 eDNA Assay Design and Validation
Robust assays for detecting eDNA are a keystone for environmental surveys and risk assessments. There is an inherent expectation that the results of targeted assays provide a high level of performance and reliability. To attain this, validation of a targeted eDNA assay requires clarity on what constitutes acceptance criteria of key factors defining assay specificity, sensitivity, and quantitative assay performance (Klymus et al. 2020; Lesperance et al. 2021). At a fundamental level, these factors need to be defined in a way that facilitates comparisons between studies and laboratories (Sivaganesan et al. 2022).
To accomplish qPCR-based assay design and validation, a general workflow includes identifying phylogenies of target and confounding taxa, sequencing and alignment of mitochondrial DNA sequences, primer and probe design, systematic specificity validation using voucher specimens, sensitivity determination with synthetic DNA, and field validation of sites with known target species' occupation and negative controls (Langlois et al. 2021). Herein, we feature the use of whole mitochondrial genome (mitogenome) sequences within this workflow and put out a call for the augmentation of these valuable resources. To facilitate ease of use, our design approach features having the same stringent amplification conditions (where possible) to allow assay runs targeting different species on the same plate (thus, using the same PCR conditions) to accommodate broad stakeholder needs.
2.1 Specificity
The development and validation of a targeted eDNA assay requires steps for determining and reporting assay specificity (Thalinger et al. 2021). These steps serve to establish the assay's ability to distinguish target DNA sequences from confounding non-target (e.g., sympatric/allopatric) organism sequences to ensure that there is a reasonable expectation of the assay to detect target taxon DNA to the exclusion of non-target taxa within an eDNA survey. Gathering natural history information of target and possible confounder populations was accomplished through early communication between field and lab practitioners involved in specific surveys. To effectively accomplish this, access to appropriate DNA sequences from target and non-target taxa is critical. Ideally, this is in the form of complete mitogenome sequences from multiple individuals per taxon from diverse geographic locations to provide the maximal amount of sequence from which to select the best DNA sequences that satisfy the requirements for specificity and sensitivity of eDNA assays (Langlois et al. 2021). Public databases such as the National Center for Biotechnology Information (NCBI) GenBank are invaluable resources for obtaining the required sequence information. In preparation for multi-mitogenome alignments as described below, we downloaded and utilized thousands of sequences from the NCBI database. For example, for the eFish1 assay, we used 4442 mitogenomes—4211 fish and 231 non-fish.
Despite the apparent high number of mitogenomes that were used, there are still substantial gaps in mitogenome sequence resources. Throughout the iTrackDNA project, we successfully sequenced and assembled the complete mitochondrial genomes (mitogenomes) of several species of interest using mtGrasp (Lopez et al. 2025). This significantly enhanced flexibility in identifying target regions for assay design, moving beyond the constraints of limited metabarcoding regions. Despite an increase in full mitogenome sequences, there are still many species that are not directly represented, and intraspecies or regional variation is still not yet adequately captured, as access to appropriate voucher specimen DNA is a limiting factor.
To address this, cost-effective whole genome shotgun sequencing of genomic DNA (gDNA) from voucher specimens and mitogenome assembly are facilitated through genome skimming using 150 bp paired-end Illumina NovaSeq 6000 reads, e.g., (Hoban et al. 2022; Lopez et al. 2025). Curated collections from museums and institutions are invaluable sources of material, and the application of whole genome amplification techniques, such as through GoEnrich (Lopez et al. 2024), can faithfully augment sparse gDNA resources for sequencing and subsequent eDNA validation. Using GoEnrich and mtGrasp, we assembled ~400 mitogenome sequences from voucher specimens representing ~120 species. These sequences have been deposited in GenBank (BioProject PRJNA1139757) to fill gaps in sequence database knowledge. An example of the impact of this new knowledge is in iterative field use and further gDNA validation of the eFish1 assay that was designed to broadly detect bony fish. Initial designs with available mitogenomes revealed a broad fish assay, but novel mitogenome alignments and voucher specimen gDNA validation indicated that DNA from some amphibian species (Ascaphus, some Ambystoma and Spea) could also be detected (Files S1 and S2). This should be considered when performing field surveys.
2.1.1 In Silico Screening Against Target and Confounding Non-Target Taxa
The first step in our workflow involves creating alignments of mitogenome DNA sequences of the target species to available confounding non-target taxa using MAFFT (Katoh et al. 2019) with default settings and visualized using Geneious Prime (Biomatters Ltd., Aukland, New Zealand) as described previously (Allison et al. 2023). Consistent with the CSA standard (Abbott et al. 2023), we paid particular attention to taxa that are closely related to the target taxa that may confound the results. Additionally, as human, domestic dog, and domestic cat DNA ubiquitously contaminate environmental samples (Clare et al. 2022), we routinely included these species in DNA sequence alignments. Moreover, we considered additional non-target organisms' DNA that the practitioner has significant contact with (e.g., mice, hamsters, goldfish, cattle, etc.), when relevant, as a possible cofounder to ensure that the chosen sequences are unlikely to generate a false positive signal.
We found that querying complete mitogenome sequences with unikseq (Allison et al. 2023) was beneficial in identifying DNA regions possessing unique characteristics in an unbiased way from which primers and probes may be selected for testing. In contrast to the propensity for eDNA assay design to historically be based on the cytochrome oxidase 1 (co1) gene, we found that mt-co1 was not a commonly chosen gene from which to derive fully validated eDNA assays (Figure 1). Rather, the mitogenome regions that supported successful eDNA assay designs spanned 18 genes overall (n = 125; Figure 1A). The most represented genic regions were mt-cyb, mt-nd1, mt-nd2, and mt-nd4 (n = 125; Figure 1A). The most represented genic regions by animal group were: mt-nd1 and mt-nd2 (n = 59; fish), mt-cyb and mt-nd5 (n = 25; mammal), mt-co1 (n = 18; invertebrates), and mt-cyb (n = 21; herptile) (Figure 1B–E). The number of bird assays designed was too low to make a general statement (n = 2; Figure 1F).

Once unique DNA regions were identified by unikseq, we used the Beacon Designer (PREMIER Biosoft, California, USA) primer/probe design software on those regions to identify the best forward/reverse primer-probe combination for in vitro screening based upon the guiding principles for primer/probe selection defined by the software (Table 1). The primers/probe candidates that have desirable characteristics were then examined in Geneious Prime with respect to the alignments to ensure that they were sufficiently different from DNA sequences of non-target taxa (Figure 2 and File S1). Any overlapping sequence identity to non-target taxa was identified, and assay designs were iteratively evaluated to obtain the most favorable assay performance characteristics.
| Primer design |
|
|
|
|
|
|
|
|
|
| Probe design |
|
|
|
|
|
|
|
|
|
| Advanced design settings (primer set and probe combination) |
|
|
|
|
|
|
|
|
|
|
|
|
|

To aid in determining assay component specificity in silico, we also used NCBI's Basic Local Alignment Search Tool (BLAST) for short sequences against the non-redundant (nr) nucleotide database. While this was a powerful tool, we realized the output only shows the first 100 results according to the level of different proxies (e.g., coverage, percent identity, E value). If the values of these proxies are equal, then the results are presented in random order. If the percent sequence identity remains high in those results, there are likely other sequences with high sequence identity that did not make the list and, hence, may be missed in the specificity evaluation. For species-specific and animal group (e.g., Cervus spp., Odocoileus spp., Salmonids, or bony fish) assays, this is particularly important when evaluating cross-hybridization from common contaminating DNA such as human, cat, and dog, or other relevant confounders. When this occurred, we queried each primer and probe candidate sequence against narrower categories (e.g., human, bony fish, etc.) rather than the whole NR database.
2.1.2 In Vitro Screening Against Target and Confounding Non-Target Taxa
While the final eDNA assay will always be comprised of a primer pair and hydrolysis probe, both SYBR and Taqman chemistry are used at this empirical validation stage; the former for initially testing the primer sets, and the latter for testing promising primer sets in combination with a hydrolysis probe containing a fluorescence dye such as FAM at the 5′ end and a quencher (e.g., ZEN/Iowa Black FQ) at the 3′ end. This two-phase approach is cost-effective to evaluate different designed assays as primers are much cheaper than hydrolysis probes and allows for more powerful troubleshooting during assay design.
We generally first test at least four technical replicates for each gDNA sample from target and non-target taxa voucher specimens and, once we establish that the desired amplification was achieved (see below), then we expanded the evaluation to between 12 and 25 technical replicates. This approach is more stringent than the minimum of four required by the CSA standard (Abbott et al. 2023). We adopted this approach to more effectively determine off-target detections.
We routinely normalize the gDNA samples to a low concentration (e.g., 10 pg/reaction). A typical 15 μL reaction contains 2 μL (10 pg) gDNA that is expected to reliably produce a detection in all technical replicates (e.g., a cycle threshold (Cq) value < 30) using the run conditions indicated in Table 2 on a Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories (Canada) Ltd., Mississauga, Ontario). While not presented herein, other thermocyclers have been used and assays tested in multiple labs with similar results. While a number of enzymes and reaction mixes could be used, we used the QIAcuity EG PCR [Cat. 250111, Qiagen, Hilden, Germany] or SensiFAST SYBR [Cat. BIO-98020, FroggaBio Scientific Solutions, Buffalo, New York, USA] kits for the SYBR reactions and the QIAcuity Probe PCR [Cat. 250101, QIAgen, Hilden, Germany] or Immolase [Cat. BIO-21048, FroggaBio] for the Taqman probe-containing reactions. Further information on the enzymes and reaction conditions has been published in detail previously (Acharya-Patel, Groenwold, Allison, and Helbing 2024b; Acharya-Patel, Groenwold, Lemay, et al. 2024a).
| Thermocycle temperature sequence (°C) | Time sequence (s) | Cycles | MgCl2 (mM) | Reaction volume |
|---|---|---|---|---|
| 95, 64a, 72 | 15, 15, 30 | 50 | 3 | 2 μL input in 15 μL reaction |
- a Five eDNA assays require a modified annealing temperature of 62°C. See Table 4 for details.
As intraspecies sequence variation can exist (Bernatchez et al. 2024), we attempted to evaluate at least five different individuals from the target taxon, preferably from different relevant geographic locations. However, this often was not possible due to limited availability of voucher specimen material. For non-target taxa, at least one individual of available confounding taxa was included when possible. There should not be any amplification of non-target taxa DNA observed within 50 cycles. In some cases, non-target taxon gDNA did produce hits, and these were recorded, and the assays were assessed to determine if those assay performance caveats were appropriate for the intended use. We routinely used eight technical replicates for the no template controls (NTCs), and all assays returned no observed detection within 50 cycles. The taxa tested and the results of the specificity testing were recorded in a technical bulletin for each assay (Figure 3 and File S2) to ensure that relevant performance criteria are consistently recorded and communicated between eDNA practitioners and end-users. The technical bulletin provides an accessible way for practitioners to provide information regarding the extent of validation of a given assay, irrespective of its origin, that can be provided in reports while supporting the ongoing process of assay validation. When some relevant species are absent from the bulletin, this should prompt the user to ensure that the assay is tested on voucher specimens of the missing species.
While the use of a hydrolysis probe provides an essential assay component to ensure specificity of a robust eDNA assay (Abbott et al. 2023), further verification of the detection of the correct DNA template can be accomplished through multiple means. In some cases, if the amplicon was large enough (e.g., ~> 200 bp), we confirmed the amplicon sequence through Sanger sequencing. Additionally, we routinely performed melting curve analysis to establish that primer dimerization was not occurring, and agarose gel electrophoresis provided an indication regarding the number and sizes of bands generated during amplification. The checklists that we routinely used for phase 1 (SYBR) and phase 2 (Taqman) validation performance are provided in Table 3.
| Phase 1 | Phase 2 |
|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
- Note: Each reaction contained 10 pg voucher specimen gDNA.
Given the complex and varied nature of environmental samples, we have elected to apply a high level of stringency to mitigate false positives. Stringency refers to the strictness with which Watson-Crick-Franklin base pairing is required for annealing to their complementary target DNA sequence. Stringency can be most easily influenced by annealing temperature and magnesium concentration, with lower annealing temperatures or higher magnesium concentrations each reducing stringency. We chose to routinely utilize a higher stringency annealing temperature of 64°C rather than the less stringent 60°C that is often the default recommendation and keep the magnesium concentration constant (3 mM MgCl2 (Table 2)), as temperature manipulation is easier to accomplish and the assays demonstrated strong performance at the higher annealing temperature (Acharya-Patel, Groenwold, Allison, and Helbing 2024b; Acharya-Patel, Groenwold, Lemay, et al. 2024a). We also chose to avoid using minor groove binder (MGB) probes in preference to maximizing sequence selectivity within the assay, as MGB probes require shorter probe designs to maintain a Tm similar to regular probes (Kutyavin et al. 2000). This approach was successful for this research project (Table 4).
| Scientific name | Common name | Assay code | Genea | Forward sequence (5′-3′) | Reverse sequence (5′-3′) | Probe sequence—IDT 5′ FAM with ZEN quencher (5′-3′) | References |
|---|---|---|---|---|---|---|---|
| Accipiter gentilis | Northern goshawk | eACGE3 | mt-nd4 | TGGTGACTTTTAGCAAAC | GGTAGATCAGTTGAATAGTG | CTCTCCCACCAACTACAAACCTGA | (Lopez et al. 2024) |
| Acipenser fulvescens | Lake sturgeon | eACFU1 | mt-nd1 | TGGCAATTTCACTCTACAC | TTGGCGTATTCAGCTAAG | AACTGGTCTCCGGCTTCAACGTAGAATATG | The present study |
| Acipenser medirostris | Green sturgeon | eACME3 | mt-d-loop | GGAACATTTATAACCTGCC | GAGTCATGTGAGGGTAAG | ATGGTACAATGACATATCCTTGACGCTA | The present study |
| Acipenser transmontanus* | White sturgeon | eACTR2b | mt-d-loop | CTACCATTAAATGTTATACACC | CCCTCATCTCATTAACTTTA | CATATCTCCATGTTCGGCTCTTACTAC | The present study |
| Acipenser transmontanus*† | White sturgeon | eACTR4 | mt-d-loop | GGTACAATGACATATCCC | CCATGATTGTCATGTGAA | AAACCAGGCTCTGTGAAACATCC | The present study |
| Alces alces | Moose | ma-eALAL4 | mt-nd3 | GGCACCATACTCACTATG | GGCATAAGAGAGAGGATATG | TATTAGCTGCAAGCCTAGCCTATGAAT | The present study |
| Ambystoma macrodactylum | Long toed salamander | eAMMA3 | mt-nd2 | CACTCTCTTCACTATTAAGTTTA | GGTTGTTTTGTCGTCAAA | CATACTCAATTTCTCTTACAACTTCACCCA | The present study |
| Ambystoma mavortium † | Western tiger salamander | eAMMV4 | mt-d-loop | TATCTGCCACACGACTATTT | AATTGATGAATGAATCTTCG | CGCCACTGGTTAAAATCTATGGAC | The present study |
| Ammodytes personatus | Pacific sand lance | eAMPE5 | mt-rnr2 | AGACGCCAAGACAGACC | CTGTTCATTAGAGCTGTAGC | CCATAAGGAGCCCAAACCAAATGGT | (Robinson et al. 2022) |
| Anaxyrus (Bufo) boreas | Western toad | eANBO5 | mt-nd1 | TACCCATCTCACTCTCTGAC | CTCAGATTCTCCTTCAGTAAGG | CAGACAATTTCTTATGAAGTCACCCTG | (Acharya-Patel, Groenwold, Allison, and Helbing 2024b; Acharya-Patel, Groenwold, Lemay, et al. 2024a) |
| Anguilla rostrata # | American eel | eANRO1 | mt-cyb | CATCTCAACTGCCTTCTCC | ATGGAGGAATAGGAGGTGA | CAACCTACTATCTGCCGTTCCATAC | The present study |
| Anoplopoma fimbriatum | Sablefish, black cod | eANFI6 | mt-nd2 | AGCAGCAATACTTCTCTT | CAAGGTGATTAAGGTGATG | ACTACTAATGCGTGACTAACAGGACAATG | (Acharya-Patel, Groenwold, Allison, and Helbing 2024b; Acharya-Patel, Groenwold, Lemay, et al. 2024a) |
| Ascaphus montanus | Rocky Mountain tailed frog | eASMO9 | mt-cyb | ACTTTATTACGGCTCTTACTTG | GTACGTTTCCGATGTAAGGGA | ATACGTATTACCATGAGGACAAATATC | (Veldhoen et al. 2016; Hobbs et al. 2019; Klymus et al. 2020; Lesperance et al. 2021) |
| Ascaphus truei | Pacific (coastal) tailed frog | eASTR4 | mt-cyb | GAACATTGGCATTATCCTACTT | AGGCGAAAAATCGTGTTAAC | CGCTTTTGTAGGGTATGTGTTACCG | (Veldhoen et al. 2016; Hobbs et al. 2019; Klymus et al. 2020; Lesperance et al. 2021) |
| Bison bison | Bison | eBIBI4 | mt-nd4 | AGCTATTTGTAATAATGTCAAC | GTATATACAGAGAGTATAGGG | CACTCCCATTAGAATAATTGTAATGTTAGA | (Lopez et al. 2024) |
| Cancer productus † | Red rock crab | eCAPR4 | mt-co1 | CGCGTATAAACAACATAAGG | GGAGGGTAGACAGTTCAG | TTCCAACACCTCTTTCTACTAATCCTCTT | The present study |
| Canis lupus #† | Wolf | eCALU1 | mt-cyb | CCTAGTATTCTCCATCCTAATC | GGCCGGAATATTATGCTG | TGGCATTCATTCCACTCCTCCA | The present study |
| Carassius auratus | Goldfish | eCAAU5 | mt-cyb | CTATACATTACACCTCAGACA | GAATTAGTCAGCCGTAGT | TTCAACCGCATTCTCATCCGTTACC | The present study |
| Carcharodon carcharias | Great white shark | echCACA2 | mt-nd5 | CCGAGTAGGAGATATTGG | GGTAGGGTTAAGTTTATGTC | CTAATTCTTAGCATAGCTTGACTGGCT | The present study |
| Carcinus meanus | European green crab | earCAMA1 | mt-co2 | ATGGCAACTTGATCTTATC | GCACATCATATAACCTACAA | CGGATTCCAGGATAGTGCTTCTC | The present study |
| Castor canadensis | Beaver | emaCACA3 | mt-co1 | GCAGTCCTTTTACTACTC | CGAAGAAAGTGGTGTTTA | CCTTACCAGTCCTAGCAGCCG | The present study |
| Catostomus catostomus | Longnose sucker | eCACA2 | mt-cyb | GTAATAATGACTGCCTTCGTAG | GAGGTGGATGATAGTTGCT | CTTATCCGCAGTTCCCTATGTAGGA | The present study |
| Catostomus commersonii | White sucker | eCACO4 | mt-nd2 | CTAACTCTCCTTGCCCTA | TGGTAGTAACTATAATTGGACTT | TTCATAACATCAGCAGCATTCCTTACACT | The present study |
| Catostomus macrocheilus | Largescale sucker | eCAMA2 | mt-nd2 | AGAGCCCCATTATAGTCA | GCAGAATAAGTCACTTAGGTA | ACTGCCTTGGTCTTACTATCTCTTGG | The present study |
| Cervus spp. | Cervus species | eCervus3 | mt-co1 | ATATAAACCTCTGTCTTTAGATT | ACGAATCAGTAGGCTTAA | TACTCTGTATCTATTATTTGGTGCCTGAGC | The present study |
| Chelydra serpentina | Snapping Turtle | eCHSE1 | mt-nd6 | CCACTAAAATCAAAACGAATA | GAGCATGAAGTGTAGAAG | CCATCACCATCAACTGTAGCACAAC | The present study |
| Clemmys guttata | Spotted turtle | eCLGU3 | mt-cyb | ACTTCTTACTACCGTTCAC | TCACTCAGGCTTGATATG | CCTAACAATAATACACCTGCTCTTCCTACA | The present study |
| Clupea pallasii | Pacific herring | eCLPA1 | mt-rnr2 | TTACTACTGAGCCATCCTA | CCGATCCAACTTACACTT | AGTTCATTCTTCTTGTTACTCGTTCTAGCA | The present study |
| Contia tenuis | Sharp-tailed snake | eCOTE3 | mt-nd4 | CACATAGGCTTAGTCATTGC | TTATTAGGCTGGTTAGGAGTC | CTCCTCAGCACTCTTCTGCTTAGCCAACAC | (Matthias et al. 2021) |
| Corbicula fluminea | Asian clam | eCOFL13 | mt-rnr2 | GATCTGAAATGGTTTAACGG | CCCCAATTAAAGGGCAAA | TGCTACCTTAGTACGGTTTCAATCGC | The present study |
| Coregonus artedi | Cisco/Tullibee | eCOAR7 | mt-nd6 | CACCACAAATAGCGTTAG | GTAGCCCTAATATACTCTTCA | CACACACCACCAACAGTCCC | (Lopez et al. 2023) |
| Coregonus clupeaformis | Lake whitefish | eCOCL1 | mt-nd2 | CATCATTCCTCTCATAGCA | ATTGGGTGGGTTAATTGT | CCATTCTCCAACCAGTCAAGCATTAGT | (Lopez et al. 2023) |
| Coregonus zenithicus | Shortjaw cisco | eCOZE1 | mt-co1 | ACGGGCATTGTCCTAGCAAA | TGCTGTGAAGAGTGTATCCGG | TCGATAGGGGCTGTTTTCGC | The present study |
| Cottus cognatus | Slimy sculpin | eCOCO3 | mt-nd1 | CTCCTGGCTCCAATTCTA | GGGCGAGAATAAACAAAATAC | ACCCTTGCCCTCACTCTATGG | The present study |
| Crassostrea gigas | Pacific oyster | eCRGI2 | mt-tl1 | CCAAGAAATTGGCCTTTA | GCTGAATTAACTTTAATAATTTAAG | CTAAGCCAACACCAGTATTCTCATG | The present study |
| Crotalus oreganus † | Northern Pacific (western) rattlesnake | eCROR1 | mt-cyb | TCCTCTATTACACACATCTCC | GGATGTGAAGTGAGGAGATAG | CATCTATTACGGCTCCTACCTAAAC | The present study |
| Ctenopharyngodon idella | Asian grass carp | eCTID1 | mt-nd4 | CCTACTCCAACAATCTACA | CTAGGAATGCGATTAAGC | AATTCAATACTCACAACCACTACTCCTAGA | The present study |
| Culaea inconstans | Brook stickleback | eCUIN2 | mt-nd2 | CCTAGCTTACTCTTCAGTC | GTGGCTAGTGAATTAATAGTG | CCACCTCGGCTGAATAACACTAAT | The present study |
| Decamastus gracilis | Decamastus | eDEGR2 | mt-nd4 | CGCCTATCATCCTATTCTG | CTCAACAACGACACCATC | CGAACCAGCAGACTACACAGATACAC | (Acharya-Patel et al. 2025) |
| Dreissena bugensis | Quagga mussel | eDRBU5 | mt-rnr1 | CGGTTAAACCGATGTTCC | CTCCTATGCGGAGACTAC | TTTCTTTCTTAACATACCGCCGTTGAC | The present study |
| Dreissena polymorpha | Zebra mussel | eDRPO5 | mt-tm | GAAGGGTGATTTACACAC | GCAACAAACTATAAGGGTTA | TTCAGGGTATGAGCCTAACAACCTTT | The present study |
| Dreissena spp. | Zebra/quagga mussel | eDRspp7 | mt-rnr1 | GCGGGGACTATGAACAAAACG | GGGTAAATTGTCAACGGCGG | AAAAACCAAGAGGCTTGGCG | The present study |
| Emydoidea blandingii | Blanding's turtle | eEMBL4 | mt-nd5 | AGGCTTCATCAAAATCTATA | CACAGGTAAAAGTTCTTTG | ATAACCTCATTCCTCCTAACACTACTACT | The present study |
| Engraulis mordax | Northern anchovy | eENMO2 | mt-co2 | AGGCATGATACTGTCATTA | GAGAGTTGGAATAAGCAC | TTATCGCCTTGTCACTCTGGTCCTT | The present study |
| Enhydra lutris | Sea otter | eENLU6 | mt-cyb | TTCATCATTTCAGCACTG | GGATAGGATTAAGGCTAGTA | ATAATCCACCTCCTATTCCTCCACGA | The present study |
| Esox americanus | American pickerel | eESAM2 | mt-d-loop | CTAGGCGATTCTTAATATCC | GTAGGGCTGAATTTAACAA | ACCCCAAAACCAGGACTATCACTT | The present study |
| Esox americanus vermiculatus | Grass pickerel | eESAMve1 | mt-d-loop | TGCTCTACTTGTATCTATACTA | GATGGATAATTGATGGGAAA | AGCTCGCATCACTAACCTCAA | The present study |
| Esox lucius | Northern pike | eESLU1 | mt-cyb | TCTCCACAGCCTTCTCATC | CCGCCTCAGATTCATTGG | CTCCTCCTAACAATAATAACCGCCTTCGT | (Lopez et al. 2023) |
| Esox niger † | Chain pickerel | eESNI3 | mt-nd2 | ACAGGTGAATGAGATATTAAC | AGGTGGATAAAATAAGTCCA | CGTGACTTCCTCAATGATTCTACTAGC | The present study |
| Eubranchipus oregonus | Oregon fairy shrimp | eEUOR1 | mt-co1 | GGACTGGATGAACAGTAT | AATAGAAGATACACCTGCTAA | CTCCACTATCTTCAGGTATTGCTCACG | The present study |
| Exoglossum maxillingua | Cutlip minnow | eEXMA1 | mt-nd5 | CACCTGTCATAACAATACC | GCTGATTGTCCTAGAATTAG | CTCTTATCGTGACTATTCTTGGACTTCTGA | The present study |
| Fish#† | Bony fish | eFish1 | mt-rnr1 | CACCTAGAGGAGCCTGTTCTA | CTACACCTCGACCTGACGTT | TATATACCRCCGTCGTCAGCTTACCC | (Klymus et al. 2020) |
| Glycera nana | Glycera | eGLNA3 | mt-nd6 | ACTGCTATTCAACCAAAC | GAAGCGTAATTGTTGTTATAG | TCCTGACGCCTACAATCGCC | (Acharya-Patel et al. 2025) |
| Glyptemys insculpta* | Wood turtle | eGLIN1 | mt-cyb | GCTCTTCCTACACGAAACTG | GATGTATGTAGGGTGGGTATC | CTCATCTTAATATTAACCTTCCTACT | (Acharya-Patel, Groenwold, Allison, and Helbing 2024b; Acharya-Patel, Groenwold, Lemay, et al. 2024a) |
| Haliaeetus leucocephalus | Bald eagle | eHALE3 | mt-nd2 | GGAGGATGAATAGGCTTA | GACATATAAGTAGAAGGTTAGAA | TAGACGATTACTGCGGTTATCCATCC | (Lopez et al. 2024) |
| Heteromastus filobranchus | Heteromastus | eHEFI7 | mt-cyb | CGGACTTCATGTAATTATCG | TATTCTTACTTCAGGCACATA | AACCTATTCCGAACTTATTGCTGCCAT | (Acharya-Patel et al. 2025) |
| Hiodon alosoides | Goldeye | eHIAL6 | mt-nd2 | CCTGTACACTTCTGGATA | GTGTGAGATTTGGCATAA | TACTACAAGGCTTAGACCTAACCACAG | (Lopez et al. 2024) |
| Hippoglossus stenolepis | Pacific halibut | eHIST2 | mt-rnr2 | TTTCAAGCAAAGAGAACCTA | GTGCATGAGTGGAGAATC | CTTCTATTCAATTCACAGGCAACCAGC | The present study |
| Holbrookia species † | Spot-tailed lizard | eHolbrookia1 | mt-nd1 | CGCACACTACTACTTACA | GGTCTTACAGGAGCTAG | AACACTACCAGCATCACTACTAATACTCC | The present study |
| Hypomesus pretiosus | Surf smelt | eHYPR4 | mt-co1 | AGCTTCCGTAGATTTGACC | CCGAAGAATCAGAATAGATGC | CCATCTTGCAGGAATCTCCTCTATT | (Robinson et al. 2022) |
| IntegritE-DNA | Endogenous DNA | ePlant5 | cl-23S | TCTAGGGATAACAGGCTGAT | TGAACCCAGCTCACGTAC | TTTGGCACCTCGATGTCGG | (Veldhoen et al. 2016; Hobbs et al. 2019) |
| Lepus americanus | Snowshoe hare | eLEAM1 | mt-nd5 | CCTCATTTCTAATCTTATCTCA | GTTGGTTTAGTTCTATGGC | ATCCCTCAAATAACCATACCCTTATACAT | (Lopez et al. 2024, 2025) |
| Lithobates (Rana) catesbeiana | North American bullfrog | eLICA5 | mt-nd5 | ATACACCGCACTATTACT | AAGAGGACTGATAGGTAAG | CCTTAACTCAGCCTGACAACTTATT | (Allison et al. 2023) |
| Lithobates (Rana) pipiens | Northern leopard frog | eLIPI1 | mt-rnr1 | AGCTTACCATGTGAACGTCTT | TACTACTAAATCCACCTTCGCT | CAATTGGCTACAATTTCTAATATAGAACAA | (Klymus et al. 2020) |
| Lota lota | Burbot | eLOLO4 | mt-nd4 | CTTGCTGCTGTATTACTAA | ATAAACTATTTCCTTGGAGAG | ATGAGTCGTATTATACCATAACCGCCTAGT | (Allison et al. 2023) |
| Metacarcinus gracilis # | Graceful rock crab | eMEGR3 | mt-co1 | ACTCCTGCTAAGTGTAAG | CCTCCTTAACATTACTCCTTA | ATCAACTGAGGCTCCAGCGT | The present study |
| Metacarcinus magister | Dungeness crab | eMEMA3 | mt-co1 | CGGGTCATCTTTTATTAACTC | GAGAGAAAATTAAAATGACTAGAA | AGGAAACACCGGAGCCTCTC | The present study |
| Micropterus dolomieu | Smallmouth bass | eMIDO1 | mt-cyb | CAACTACGGCTGGTTAATC | AGCGTCAGAGTTCAATCCT | AACCTCCTCTCAGCCGTTCCTTACATT | (Klymus et al. 2020) |
| Micropterus salmoides | Largemouth bass | eMISA2 | mt-co1 | CCTACTCCTTCTGTCACTTC | TCATGGCTCAAACTATACCC | TCCTTACAGACCGAAACCTCAACAC | (Klymus et al. 2020) |
| Morone saxatilis | Striped bass | eMOSA1 | mt-atp6 | CGCTTACTTGTCCTTCAA | CAAGCATATTTAGGGTAATAAGAA | TTTACACAACAACTGCTTCTTCCCTT | The present study |
| Myotis spp. | Myotis bats | eMyotis1 | mt-rnr1 | TTCGCCAGAGTACTACTA | GCTGATTTAGCAAAGATTG | TCGATTATAGAACAGGCTCCTCT | The present study |
| Myotis lucifugus | Little brown myotis | eMYLU4 | mt-nd5 | CCTACTCATCCGATTCTAC | GGTGGTAATTGCTCCTAG | AGAGCAATAAAATAATCCAATCCTTAGCCC | The present study |
| Myotis septentrionalis | Northern myotis | eMYSE3 | mt-co2 | CGAACTAACAATCCGTATG | CAGTGTTGTCTGATTTAGG | TATTGCACTCCTGAGCTGTTCCTTC | The present study |
| Myotis yumanensis | Yuma myotis | eMYYU7 | mt-nd4 | ATCGCATACTCATCTGTTAG | CCATGAAACTTCAGGGAC | TGAATTAAGACAGCCATAATCACAAGCG | The present study |
| Odocoileus spp.*† | Odocoileus species | eOdocoileus2 | mt-co1 | ACAGGAGTAAAGGTCTTTA | CGTAGTAAGTATCGTGAAG | TGGAGGCAACATTAAATGATCACCT | The present study |
| Oncorhynchus clarkii † | Cutthroat trout | eONCL4 | mt-nd1 | TTGCACTATCCAGCCTAGCT | GGCTGGTACGAGCAGTCAA | TACAGGAGGATTTACTCTTCAAACCTTCAA | The present study |
| Oncorhynchus gorbuscha | Pink salmon | eONGO5 | mt-nd1 | CAAGCCTGGCAGTATATTCC | ACCAGTAGTCAGATGCTTTCC | CATTATCACAGGAGGATTCACTCTTCAAAC | (Su et al. 2023) |
| Oncorhynchus keta | Chum salmon | eONKE9 | mt-nd4 | GCGAAATGGCTATGAACC | CGATCACTTAAGTCAGGTTA | TCAAAGTCTAATTATCGCCCTAGCAAGT | The present study |
| Oncorhynchus kisutch | Coho salmon | eONKI4 | mt-nd1 | CCCTTTTGACCTCACAGAG | GGTTCAGGGCAGTTAGTTCT | TGCCCTCTTTTTCCTAGCCGAGTACG | (Klymus et al. 2020; Lesperance et al. 2021; Hocking et al. 2022) |
| Oncorhynchus mykiss | Rainbow (steelhead) trout | eONMY5 | mt-nd2 | GTCTCTCCCTGTATATCG | GAGTATTGATGGTTAAAGAGT | TGACATCTTCAGCCTTCCTCACAT | The present study |
| Oncorhynchus nerka | Sockeye salmon | eONNE2 | mt-cyb | TGAGGACAAATATCCTTCTGG | GCAATGACGAAAGGGAAC | TCCGTACGTCGGGGGCGCC | (Su et al. 2023) |
| Oncorhynchus tschawytscha | Chinook salmon | eONTS5 | mt-nd1 | ACTATTTATTAAAGAACCCGTC | GCTCAGCCTGATCCCAA | TCCTGTTACAGATCTTAACCTAGGGGTAC | (Su et al. 2023) |
| Ophelina acuminata | Ophelina | eOPAC1 | mt-nd2 | CAACCATATAGGATGAATACTA | GAGGATAAGGATTGAGGA | TCGTCATCCGCCATTGCCTA | (Acharya-Patel et al. 2025) |
| Oreamnos americanus # | Mountain goat | eORAM2 | mt-co3 | CCTTCACAATCTCAGATG | CAGACGATTAAGAAGGTG | TTCTTCGTAGCCACAGGATTCCA | The present study |
| Ostrea lurida | Olympia oyster | eOSLU2 | mt-co3 | CAGTCTTCTAATTTTACGCATA | TTCATCCATACACAACCATA | CACTCTTCTCATATACTTCCTCGTTCTCCT | The present study |
| Ovis canadensis | Bighorn sheep | eOVCA1 | mt-d-loop | TCTGCTAAAACTTCCAAAC | ACGTGTTCATACATTCAAG | TTCCACTCCACAAGCTCACAC | The present study |
| Pantherophis vulpinus (P. gloydi) | Eastern foxsnake | ePAVU5 | mt-nd4 | AAACCCGAATTATAATCCTTAC | TTGGACACCAATTAAATAGTG | CCTGCCGATACTAACAACCTGATGG | The present study |
| Parelaphostrongylus tenuis | Brainworm | ePATE1 | mt-co1 | GGGACTAGATGAACTGTG | CATAAAATTAATACCGCCCAG | TCTTCTACCAACATGACCTAAAGTACTCAA | The present study |
| Pekania pennanti | Fisher | ePEPE2 | mt-cyb | CCTACTTCACACCTCAAA | GCCAATGATAATGAATGG | TTCCACTGGTTGTCCTCCGATT | The present study |
| Perca flavescens | Yellow perch | ePEFL1 | mt-nd5 | CTCCCATGTTAAAACTGC | TCAGGTTCAGGAAGTTAC | TTCTTAGTCAGCCTTCTTCCGTTATT | The present study |
| Percopsis omiscomaycus | Trout Perch | ePEOM6 | mt-nd2 | CCTACTTACATACTTCATCATAA | GGATATTGTTGTAGAGATTGT | TTCAAGACCAACAACACAACAAGCATTA | The present study |
| Pimephales promelas | Fathead minnow | ePIPR2 | mt-nd2 | GAGGCGGATTAAACCAAA | GCAGATGTTATTAGGATGTATG | ATTCTGGCTTACTCCTCAATCGC | The present study |
| Platygobio gracilis | Flathead chub | ePLGR2 | mt-nd1 | GCCTATATTGTCCCAGTC | CCAGGTTAAGGTTAATGAC | TCTTAGCAGTAGCCTTCTTGACCTTG | The present study |
| Prionospio (Minuspio) lighti | Prionospio 2 | ePRLI4 | mt-nd4 | TATACTTCAACGGCAACA | ATCTGACCCTGTAATTCG | TTGCTAAGGCTAATGCTGCTTCACA | (Acharya-Patel et al. 2025) |
| Prosopium williamsoni | Mountain whitefish | ePRWI4 | mt-nd1 | CAGAGTACTCCAACATCC | GAGGTTTATTGCCGTGAG | TACACTCTCGACCATCCTCTTCCTAG | The present study |
| Pseudacris spp.# | Boreal and spotted chorus frogs | ePSspp2 | mt-rnr2 | GAAGCGGGGATAAACTTA | TGTGGAGGTTTTATTTTACTC | ACGAGAAGACCCTATGGAGCTTTAAACTA | The present study |
| Ptychocheilus oregonensis | Northern pikeminnow | ePTOR1 | mt-nd1 | CCCTTTGCTTTATTCTTC | CGTTGTTAATTCTGGTATAC | CCCTATAAATAACACGGCGGACAG | The present study |
| Puma concolor | Cougar | ma-ePUCO13 | mt-co2 | GCCATCATCCTAATTCTC | GGGATTATGTAGGAGTCAAAG | AAGCTACGAGTACACTGACTACGAAGA | The present study |
| Rana aurora | Northern red-legged frog | eRAAU1 | mt-rnr2 | TGAAGAAGCGGGAATCAAA | GCATACAGATTTCTTGTGTGTG | TAAACTCATCATACACCTCTGTGCTC | (Klymus et al. 2020; Lesperance et al. 2021) |
| Rana cascadae | Cascades frog | eRACA2 | mt-cyb | CCTAATTGCCCAAATCGCT | CTCAAAATGACATCTGGCCC | TGGCTGGCTCCTTCGTAATTTACATG | (Klymus et al. 2020; Lesperance et al. 2021) |
| Rana luteiventris † | Columbia spotted frog | eRALU2 | mt-nd1 | TTCCTCTACCAATCCCCTAT | CCAGGAAAACAGTGCATAAG | CAGCCTAACCGTTTACACAATTTTGGG | (Klymus et al. 2020; Lesperance et al. 2021) |
| Rana pretiosa † | Oregon spotted frog | eRAPR2 | mt-cyb | GTAACCTCCATGCTAACGGT | CTATCACTAGGAATAGGAGGATC | TTTCCACATCGGCCGAGGCCTC | (Klymus et al. 2020; Lesperance et al. 2021) |
| Rangifer tarandus | Reindeer | ma-eRATA3 | mt-nd5 | CTGAGCCCTCTTAATAACA | GCGAATAGACTTCCGATTA | ATTGCCACCTCCTTCACAGC | The present study |
| Rhinichthys cataractae | Longnose (nooksack) dace | eRHCA4 | mt-nd4 | TAGCCAAAACCACATCAA | AGCCATGATTAGGAAAGC | CGGCAACGTCTTTACCTCACAC | The present study |
| Rhinichthys umatilla # | Umatilla dace | eRHUM4 | mt-d-loop | GTGTAGCATTACATGCGTATAG | TTATCCGAGGGTGGAGTC | CACACTGTGAACTATTCCTTGCATC | The present study |
| Salmonidae † | All Salmonids | te-Salmo2 | mt-tiqm | CCCTCAATTCTAGAGAGA | CTGACCTTACTAGAAAGTG | TCGAACCCATCCTCAAGAGATCAAA | The present study |
| Salmo salar | Atlantic salmon | te-eSASA2 | mt-nd1 | CCCATACTTGCCCTTACG | CGCTGAGTAAGATTAGTCCAA | CCCCATGCCCATCCCTTACCCCATTA | The present study |
| Salvelinus alpinus † | Arctic char | eSAAL1 | mt-co2 | TCCGAATCCTATATCTCATG | TCGTAATCGGTGTACTCG | CGCATCTTACTATTAAAGCAATGGGCCACC | The present study |
| Salvelinus fontinalis | Brook trout | eSAFO6 | mt-co3 | GCATCATAGAAGGAGAGC | GGCTTCGTAATACTCCAT | CGTCCAAGCTCTTACTCTCACCATC | The present study |
| Salvelinus namaycush | Lake trout | eSANA1 | mt-nd2 | GGGCTTATCCTGTCTACATG | CCCAGGGATAGAAGCACTA | TGACTCTTCCTTAATTATCGCATTA | (Allison et al. 2023) |
| Sander canadensis | Sauger | eSACA1 | mt-nd2-tw | CTCTCTCTCAGCCGATAG | AGCCTAAGTCTCTTAACG | ATTCTTCTACTACCAATAACTCCAGCCATT | The present study |
| Sander vitreus | Walleye | eSAVI2 | mt-nd1 | CTCGGGATCTTGTTTCTA | CTGATACTAATTCGGATTCG | CCTATCAAGCCTAGCAGTCTACTCTATTCT | (Lopez et al. 2023) |
| Sebastes maliger | Quillback rockfish | eSEMA3 | mt-d-loop | CGAAGGTATTACATAAAGCA | GAGTGTTTGTGTTAGGTCTTA | CCAACAATCATTTATAAGGACTGAGCGAAT | (Allison et al. 2023; Acharya-Patel, Groenwold, Allison, and Helbing 2024b; Acharya-Patel, Groenwold, Lemay, et al. 2024a) |
| Sebastes paucispinus | Boccacio rockfish | eSEPA9 | mt-d-loop | CCAGTAAACAATACAAAACTAC | CCTAGAATTAAATTCAGCTTG | TACCCACCATTTCTCCAACATGCTA | (Acharya-Patel, Groenwold, Allison, and Helbing 2024b; Acharya-Patel, Groenwold, Lemay, et al. 2024a) |
| Sebastes ruberrimus | Yelloweye rockfish | eSERU5 | mt-d-loop | CTACCAGTCCACTCATAA | TGGGATTTCATATTCTACTTG | AACATAAGATATACGCCCATCAACCC | (Acharya-Patel, Groenwold, Allison, and Helbing 2024b; Acharya-Patel, Groenwold, Lemay, et al. 2024a) |
| Sorex bendirii | Pacific water/marsh shrew | eSOBE7 | mt-nd4 | CCCACTTATACTCATAGCT | CGAGTGATAATAATTAAGGTTG | ATCACCTAATACACGAGACAGCGAC | The present study |
| Sorex cinereus | Masked shrew | eSOCI1 | mt-rnr2 | TACATTCCTAATCCTTAATCC | GTTATTCGATCTGACATAAGC | CACATTAACTCCTAACCCACAACTGGACTA | The present study |
| Sorex fumeus | Smoky shrew | eSOFU1 | mt-nd1 | CCAAGAATACTCATGATTACTG | GAAATGCACCTAGAAATAGAAC | ACGTTGAATATGCCGCAGGACCATT | The present study |
| Sus scrofa #† | Wild boar | eSUSC1 | mt-cyb | ACCAGAATGATATTTCTTATTCG | AGGCTATTAGAGCTAGCA | CTACGCTATCCTACGTTCAATTCCTAATAA | The present study |
| Synaptomys cooperi | Southern bog lemming | eSYCO1 | mt-nd2 | TAACAGGACCTTTAATCACC | TGTTCATAGTCCTAGTTGC | AACTACCCTAACCCACGCTCTACAGAAG | The present study |
| Tadarida brasiliensis | Brazilian free-tailed bat | eTABR1 | mt-cyb | GCTGATTACTACGCTACCTAC | GTGATGACTGTGGCTCCT | CTATTACGGATCTTACACCTACACA | The present study |
| Thaleichthys pacificus | Eulachon/Oolichan | eTHPA6 | mt-nd1 | CTTCCTATTCCTCGCTAC | AGAACAAATAGAATCCCTAGA | CATACTAGCCCTGACACTTGCCCTC | The present study |
| Thamnophis sauritus | Eastern ribbonsnake | eTHSA2 | mt-cyb | AACTCGGAGGAACAATCG | CAATCAAGGTGAATGGTGC | CAACTCACATTCTGACTCCTAATCG | The present study |
| Thymallus arcticus | Arctic grayling | eTHAR1 | mt-rnr1 | GAGAGGCCCAAGTTGATAGT | ACCCATTTTGCTTACTATATGA | CCCCTGAACCCACGACAGCTATG | The present study |
| Thysanocardia nigra* | Segmented marine worm | eTHNI3 | mt-nd1 | ACCCTCAACATATCCTTTA | CGGAGATTAGTCTGAGAA | CACATTCCTCTTCTTATTCTCCTTCTTCCT | (Acharya-Patel et al. 2025) |
| Ursus americanus | American black bear | eURAM1 | mt-atp6 | GCCTATTCATCCAACCTA | GCTGTAATGGTGCTAATAC | CGACTAACCGCTAACATTACTGC | (Lopez et al. 2025) |
| Ursus arctos | Grizzly bear | eURAR2 | mt-nd5 | CCACAATATTACACCCACTA | ATGCCAGGATAAAACCTAA | CCCACAAACAACCATACCCTACTATCTAA | The present study |
- Note: Amplicon size and sequence information are in Table S1. *Requires a modified annealing temperature of 62°C. Some specificity limitations have been identified: #Refer to alignments in File S1 and †Refer to the technical bulletin in File S2 for details.
- a See Figure 1 legend for gene abbreviations.
- b The eACTR4 assay is the better overall performing assay. eACTR2 is shown for comparative purposes only and is not recommended for use.
Despite this, we noticed that a few assays had consistency issues in amplifying 20 copies/reaction of the appropriate synthetic double-stranded synthetic DNA (e.g., gBlocks: IDT, Iowa, USA; Strings DNA fragments: ThermoFisher Scientific, Mississauga, Ontario, Canada). Twenty copies/reaction of double-stranded synthetic DNA were routinely used as a positive plate control (see Section 4 below). We performed an eight-point thermal gradient for annealing temperatures between 60°C and 64°C (specifically 60°C, 60.3°C, 60.8°C, 61.6°C, 62.6°C, 63.4°C, 63.8°C, and 64°C) on a Bio-Rad CFX96 Real-Time PCR Detection System to determine the role of annealing temperature on the inconsistent performance. To do this, we compared the performance of assays with consistency issues with a typically robust assay (e.g., eONMY5; Figure 4). For the robust assay, amplification success remained consistent across the eight tested annealing temperatures between 60°C and 64°C. However, it is evident that the assays with inconsistent performance have difficulty producing amplicons at the 64°C annealing temperature despite the same template input amount (three examples are in Figure 4). Gradually reducing the annealing temperature reveals a common point of inflection at 62°C, under which lower annealing temperatures maintain the same strong amplification signal. Therefore, for those assays with inconsistent amplification at 64°C annealing temperature, reduction of the annealing temperature to 62°C enhanced assay performance (Table 4).
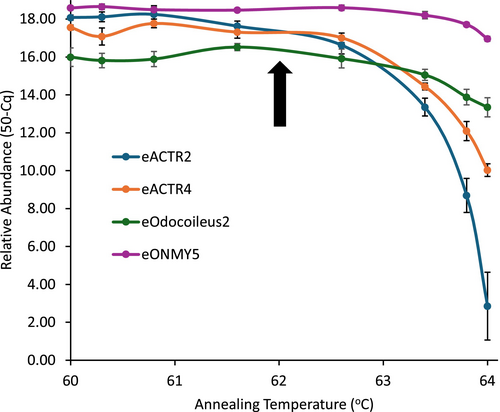
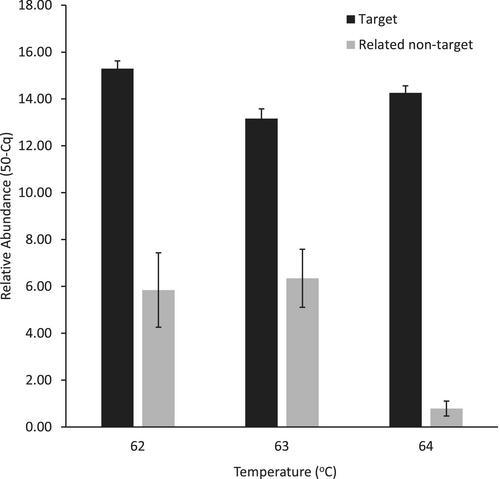
However, a reduction in stringency by decreasing the annealing temperature bears the increased risk of non-target DNA amplification. Thus, it is best that specificity is tested at the desired annealing temperature to identify any instances where a non-target taxon begins to show some hits. Generally, specificity was not compromised for the affected assays when the annealing temperature was reduced to 62°C. However, there was a notable exception—the eOdocoileus2 assay, which was designed to detect Odocoileus deer (e.g., mule and white-tailed deer) but exclude Cervus deer (e.g., elk and red deer). This was a challenging assay to design due to the high sequence identity of the mitogenomes between groups. The best DNA sequence regions were selected through unikseq and primer/probe designs were created to best satisfy the criteria of a high-performing qPCR assay. However, there was a compromise, and examination of primers and probe alignments indicates that the 3′ ends of the forward and reverse primers have sequence identity of seven and five nucleotides with Cervus sequences, respectively (Figure 2). While its amplification efficiency using a synthetic double-stranded DNA template increases from 81% to 85% to 97% for annealing temperatures of 64°C, 63°C, and 62°C, respectively, the propensity to hit closely related non-target sequences also increases at the lower annealing temperatures compared to 64°C (Figure 5). Thus, in this case, a trade-off between these performance parameters needs to be made to enhance the confidence in the use of the eDNA assay, taking into consideration the intended study location. Therefore, balancing off-target amplification with amplification efficiency, the recommended annealing temperature is 62°C for the eOdocoileus2 assay if elk are not abundant in the study region.
Despite the extensive efforts to generate/use full mitogenome sequences for assay design and empirically validate assay specificity performance in vitro with voucher specimens, there are still gaps in these important resources. In a few cases, we were unable to distinguish between closely related co-occurring species through sequence alignment and/or validation with voucher specimen DNA. These assays are marked in Table 4. This was the case for the white sturgeon (eACTR4), Western tiger salamander (eAMMV4), American eel (eANRO1), wolf (eCALU1), red rock crab (eCAPR4), chain pickerel (eESNI3), spot-tailed lizards (eHolbrookia2), graceful rock crab (eMEGR3), cutthroat trout (eONCL4), mountain goat (eORAM2), chorus frogs (ePSspp2), umatilla dace (eRHUM4), Arctic char (eSAAL1), wild boar (eSUSC1), and Oregon spotted frog (eRAPR2). In those cases, the user should determine if the assay is appropriate within their desired survey context.
In some cases, the use of a combination of assays can provide valuable insight. For example, assays to detect DNA from the at-risk Oregon spotted frog (eRAPR2) had to be designed with very limited sequence resources. While the assay had many strong characteristics, we were unable to differentiate between DNA from the Oregon spotted frog and the least concern Columbia spotted frog. Fortunately, the Columbia spotted frog assay (eRALU2) only detects its desired target and not Oregon spotted frog DNA, so a combination of the two assays' results still enables inference on whether Oregon spotted frog occupancy has occurred.
One surprise was discovering that the eFish1 assay can detect some but not all amphibians (e.g., tailed frogs, some salamanders, and toads; see Files S1 and S2). This may reveal some interesting evolutionary relationships that were not apparent before. Extensive attempts at assay redesigns have not yielded a better-performing assay. As full mitogenome resources increase, especially for amphibians, it may be possible to identify another suitable region that can distinguish bony fish DNA from other animal groups. Despite the shortcomings of the eFish1 assay, it remains a powerful tool for eDNA surveys. In many cases, gradually filling in the gaps for missing mitogenome sequences and better access to voucher specimens will enable effective assay redesign efforts to address these issues. However, in some cases, even full mitogenome sequences may not be sufficient to create an assay that is sufficiently robust, specific, and sensitive. In these situations, assay designs using nuclear genomes may provide a possible solution.
2.2 Sensitivity and Quantitative Assay Performance
Sensitivity refers to the minimum number of copies in a sample that can be measured accurately in the assay (Bustin et al. 2009). It is expressed as a limit of detection (LOD; Klymus et al. 2020; Lesperance et al. 2021), the concentration that can be detected with reasonable certainty (a 95% confidence interval is usually used; (Burns and Valdivia 2008)). Standard curves generated with synthetic double-stranded DNA can reveal the assay's sensitivity, allow the calculation of the LOD and limit of quantification (LOQ), and allow for the estimation of DNA copy numbers within a sample. These measures and associated 95% confidence intervals for an eDNA assay based on targeted qPCR are expressed in the number of copies per reaction. To generate standard curves, the CSA performance criteria require a serial dilution “of at least eight concentrations of a defined template, of which five concentrations have a detection rate of ≥ 95% and three have a detection rate of < 95%” (Abbott et al. 2023). We routinely perform a 10-point serial dilution of synthetic double stranded DNA template ranging from 62,000 to 0.032 copies/reaction, exceeding the CSA standard, as we found that this approach aptly provided the desired data coverage needed depending upon assay performance. Technical replication differed depending upon the number of copies, with higher technical replication at the lower concentrations (n = 8 for 62,000–500 copies/reaction; n = 24 for 100–0.032 copies/reaction) to ensure greater confidence in the estimation of assay performance characteristics. Consistent with the CSA standard, we also had 24 NTC technical replicates that all showed no detection (limit of blank (LOB) = 0).
While the LODs can be calculated by multiple approaches with similar results (Klymus et al. 2020; Lesperance et al. 2021), one of the robust methods for calculating the LOQ, eLowQuant, utilizes an adapted binomial-Poisson distribution (Lesperance et al. 2021). We therefore used eLowQuant to calculate both LOD and LOQ as assay performance indicators using synthetic double-stranded DNA that contains the amplicon sequence (Table S1) (Klymus et al. 2017). We chose synthetic DNA over plasmid or PCR amplicons that are also permissible according to the CSA standard, as reproducibility between batches and labs tends to be better. It should be noted that gDNA is not desirable for generating standard curves, as mitogenome copy number can vary widely between tissue types, physiological states, and individuals (Garrido-Sanz et al. 2022).
With the generated data set, two standard curves are produced: a log linear curve with continuous data for the higher DNA concentrations and a maximum likelihood curve derived from the binomial data for low DNA concentrations (Lesperance et al. 2021) (Figure 3). A LOQcontinuous defines the transition point between the two standard curve types. These, along with the LOD and LOQ, were determined and recorded along with 95% confidence intervals in the technical bulletins (File S2), satisfying the reporting criteria of the CSA standard (Gagné et al. 2021). We generated separate technical bulletins for the same assay when different enzymes (Qiacuity or Immolase) were tested. We generally found highly comparable results for these sensitivity parameters (File S2) (Acharya-Patel, Groenwold, Allison, and Helbing 2024b; Acharya-Patel, Groenwold, Lemay, et al. 2024a). Overall, for the 125 assays presented herein, the median LOD was 0.4 copies/reaction, LOQ = 1.7 copies/reaction, and the LOQcontinuous was 4 copies/reaction (n = 125) (Table 5). All CSA performance criteria were met or exceeded apart from five assays that were slightly below the 80% efficiency cut-off for continuous data (Table 5).
| Category | LODa | LOQb | LOQcontinuousc | Efficiencyd | R2 valuee |
|---|---|---|---|---|---|
| Median | 0.4 (0.3–0.8) | 1.7 (1.2–2.9) | 4 | 93% | 0.9987 |
| Maximum | 5.1 | 19.4 | 100 | 110% | 1.0000 |
| Minimum | 0 | 0.09 | 0.8 | 71% | 0.9827 |
- Note: The 95% confidence intervals for the LOD and LOQ are shown in brackets. Individual details are recorded in the technical bulletin for each assay (File S2).
- Abbreviations: LOD, limit of detection; LOQ, limit of quantification; LOQcontinuous, limit of quantification for continuous data.
- a All 125 assays exceed the CSA requirement of having a LOD ≤ 10 copies/reaction.
- b All 125 assays exceed the CSA requirement of having a LOQ ≤ 20 copies/reaction.
- c All 125 assays met or exceeded the CSA requirement of having a LOQcontinuous ≤ 100 copies/reaction.
- d Efficiency is calculated for continuous data only and is required to be between 80% and 110% to meet the CSA standard. All but five assays satisfied this requirement. eCAAU5, eEMBL4, eSalmo2, eSANA1, and eSERU5 assays, while meeting or exceeding all other performance criteria, had amplification efficiencies < 80%. See technical bulletins in File S2 for details.
- e All 125 assays exceeded the required R2 of the continuous data standard of ≥ 0.95.
3 Reporting Data
Specific considerations are required when determining the performance of assays used to detect or quantify eDNA contained within complex matrices. eDNA assay data may be reported as “detected” or “not detected” to generate qualitative results. For quantitative results, we calculated copy number from Cq values when the frequency of detection was ≥ 95% of the technical replicates using the linear regression equation of the continuous standard curve (Figure 3 and File S2). For samples with < 95% detections in a given number of technical replicates (e.g., n = 8, see rationale in Section 4), we calculated a copy number estimate with eLowQuant (Lesperance et al. 2021) along with an estimated standard error (Figure 3 and File S2). Functional examples of applying these estimation approaches on field samples can be found in the references presented in Table 4. The higher the number of technical replicates, the smaller the 95% confidence intervals and standard error estimates and the higher the chances of detection (Lesperance et al. 2021). This becomes very important to end-users if they wish to have a high confidence that an at-risk or invasive species may not be present at a given site. There are different ways to express the sample analysis results depending upon the matrix used [e.g., copies/L (water), copies/g (sediment or soil), or copies per sample (surface swabs)]. Individual assay results from field sample replicates are reported to facilitate data interpretation.
4 Routine Testing of Environmental Samples
As indicated by Thalinger et al. 2021 and in the CSA standard (Abbott et al. 2023), the highest validation level includes testing on field samples. We note that the procedure can be done in situ against well-characterized environmental samples. Moreover, eDNA samples from actual field sites known to host the target species with great abundance were tested to assess the false negative rates for many assays presented herein. Field validation is an iterative process that may require expansion depending upon survey needs, and, in some cases, finding an appropriate site may be difficult. In the same vein, field sites where the targeted taxon is known to be absent should also be harvested to establish associated false positive rates. This field validation completes the robustness of the targeted qPCR-based assay by demonstrating that it is possible to detect the target taxon DNA from field samples (Abbott et al. 2023). Ideally, field validation is incorporated within an eDNA study, wherever possible, to account for variable environmental sample and population effects. We have reported those instances where the assays have been field validated in the technical bulletins (Figure 3 and File S2), and field validation is ongoing.
Finally, the CSA standards address approaches to take to identify false negatives and positives (Gagné et al. 2021; Abbott et al. 2023). False negatives can come from failure to detect targeted species that are present on the site or because of the poor quality of the samples. False positives can come from contamination or incorrect analytical detection of non-targeted species (Rees et al. 2015; Evans et al. 2017). To avoid false positive (type 1) errors, we include field blanks in the sampling sets (e.g., one per sampling day) that are randomized along with other samples prior to sample processing (Figure 6). The number of technical replicates used for field surveys can vary from lab to lab as there may be different needs and capacities, but the minimum number of technical replicates required by the CSA standard is n = 3 (Abbott et al. 2023). However, it is important to note that the number of technical replicates chosen impacts the LOD and LOQ and their confidence intervals (Lesperance et al. 2021). We routinely use eight technical replicates (Figure 6) as there is a marked improvement on LOD and LOQ from n = 3 to n = 8, and it is a good compromise between sensitivity and cost (Lesperance et al. 2021). This decision is further supported by the eLowQuant-generated estimates of the impact of technical replicate number on LOD and LOQ shown on all technical bulletins (Figure 3 and File S2) and in previous publications appearing in Table 4.

As plate negative controls, NTCs must show no fluorescence. We routinely run n = 8 on every plate and situate the NTCs at the end of the plate immediately beside the positive plate controls (Figure 6). This is logistically easiest to set up while expecting the highest level of pipetting prowess to identify plate contamination issues. To avoid false negative (type 2) errors (Thomsen and Willerslev 2015; Evans et al. 2017), we include the required two replicates of positive controls on every 96-well qPCR plate (Figure 6). These positive controls are at a concentration expected to produce 100% detection. As suggested by the standard, we use a low copy number per reaction of the positive controls that produces reliable detection (e.g., 20 copies/reaction) to reduce the risk of contamination.
Samples can contain material that affects the DNA integrity, or the DNA may be damaged by bleach or ultraviolet light. Thus, to evaluate the probability and reduce the likelihood of a type 2 error, extracted DNA is routinely evaluated for quality and integrity (Abbott et al. 2023). The CSA standard recommends using an endogenous sample positive control (ESPC) or an internal positive control (IPC) (Abbott et al. 2023). The former is “a nucleic acid fragment that is not derived from the target taxon and expected to be ubiquitously present within an eDNA sample,” whereas the latter is “an exogenous synthetic nucleic acid fragment added to an eDNA sample either during extraction or to the extracted DNA prior to qPCR setup” (Abbott et al. 2023). Examples of published ESPC use are detection of chloroplast 16S or 23S ribosomal RNA gene sequences or using a general fish assay (Veldhoen et al. 2016; Furlan and Gleeson 2017; Hobbs et al. 2019; Zhu et al. 2024). We have elected to use a chloroplast gene-based assay as an ESPC for routine use in the context of animal taxon eDNA assays. The IntegritE-DNA assay, also known as ePlant5 (refer to Table 4; for open access details see Veldhoen et al. 2016; Hobbs et al. 2019), has been extremely useful in determining if the DNA sample is too degraded, inhibited, or otherwise compromised to affect the interpretation of the results. If a sample fails the integrity test, then we routinely subject the affected sample to an inhibitor clean-up step using a OneStep PCR Inhibitor Removal Kit (Zymo Research, Irvine, CA, USA). Often this resolves the issue if the reason for detection failure was inhibition, and the cleaned-up sample is then tested for the target DNA of interest. If the clean-up step does not result in an improved chloroplast DNA signal, then the sample is deemed poor quality, and caution is exercised in interpreting the target DNA detection result. The reader is encouraged to refer to the references in Table 4 for examples of the application of the IntegritE-DNA assay. While unable to detect sample degradation, an IPC is particularly useful for specifically determining enzyme inhibition and we refer the reader to excellent resources on this topic in the literature (Hartman et al. 2005; Goldberg et al. 2016; Hunter et al. 2019).
Another important consideration in interpreting eDNA surveys is the deposition of DNA by the target species. It is well known that organisms can vary greatly in their rate of DNA release into the environment (Allan et al. 2021). Previous studies and work herein have found that some high-quality assays developed to detect reptilian eDNA have trouble detecting their targets in environmental samples apparently for this reason (Matthias et al. 2021; Samuels et al. 2025) (e.g., spot-tailed lizard (eHolbrookia2), Northern Pacific (western) rattlesnake (eCROR1); see File S2).
5 Conclusions
Herein, we present 125 eDNA assays particularly relevant to North American species that were generated through the iTrackDNA research project that meet or exceed the recent CSA W214:21 and CSA W219:23 standards to detect target taxa by qPCR (Gagné et al. 2021; Abbott et al. 2023). These standards are increasing confidence in the use of eDNA methods, and federal and provincial regulators are incorporating them into operations and assessments. In developing the assays, we incorporated useful tools and best practices, including full mitogenome sequence evaluation where possible to maximize the likelihood of designing high-performance eDNA assays that meet reliability standards. While these assays perform well in certain contexts, it is important that users validate each individual assay on voucher specimens and field samples that are relevant to their survey needs. Recognizing that full validations of assays and high levels of technical replication may be unfeasible for resource-limited laboratories, we emphasize that our workflow seeks to fulfill best practices, and modifications can be made to prioritize the most crucial elements of assay validation prior to use. Regardless, these new eDNA tools will aid stakeholders, which include regulators, Indigenous peoples, non-governmental organizations, and those in academia and industry, to monitor biodiversity, protect ecosystems, improve environmental assessments, and ameliorate biosphere conservation.
Author Contributions
V.S.L., I.B., and C.C.H. conceived and designed the research; M.L.D.L., M.J.A., J.J.I., J.C., N.A.-P., L.C.B., M.T.B., M.-P.B., M.-L.C., L.C., A.H.D., E.T.G., H.L., I.G.M., Y.R., G.K.K., F.S., T.A.T., R.L.W., C.L.Y. acquired the data; V.S.L., M.L.D.L., M.J.A., J.J.I., J.C., N.A.-P., and C.C.H. analyzed and interpreted the data; V.S.L., M.L.D.L., J.C., J.J.I., and C.C.H. drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This project was funded through the Genome Canada's Large-Scale Applied Research Project (LSARP) program #312ITD, which also includes funding from Genome Québec and Genome British Columbia to V.S.L. and C.C.H. and the Canada's Research Chair Program to V.S.L. Additional support was from a NSERC collaborative research and development grant #CRDPJ 531846-18 to C.C.H., M.J.A. and N.A.-P. are recipients of NSERC CGS-D graduate fellowships. M.L.D.L. is a Liber Ero Fellow. The funders were not involved in the design, interpretation, or the writing of the present study. We thank Dr. Vanessa Thompson for logistical support, Dr. Anita Thambirajah for helpful discussions, Sarah Trilesky for critical reading of the manuscript, and the many participants in the iTrackDNA project for provision of samples, interlab validation, and input into pipeline refinement. Possession and disposal of wildlife tissue samples used for genetic analysis are covered under British Columbia Wildlife Act permit #NA22-782623.
The authors wish to dedicate this work to the late Dr. Louis Bernatchez, who was a scientific co-lead along with V.S.L. and C.C.H. on the iTrackDNA project.
Conflicts of Interest
C.C.H. is coinventor on U.S. patent #11,479,813 issued to the University of Victoria that includes the ePlant5 aka IntegritE-DNA assay for commercial use. Otherwise, the authors have nothing to report.
Open Research
Data Availability Statement
Raw data are available upon request to the corresponding author. Mitogenome assemblies are publicly available through the National Center for Biotechnology Information (NCBI)'s GenBank Bioproject PRJNA1139757.




