Tropical Rainfall and eDNA Washout Impact Estimations of Amazonian Biodiversity Patterns From Environmental Samples
Funding: This work was part of the IQCN, OHM-Oyapock, DEBIT (ANR-17-ce02-0007) and Vigilife Sentinel River projects. It was also supported by Investissement d'Avenir grants managed by the Agence Nationale de la Recherche (CEBA: ANR-10-LABX-25-01, DRIIHM: ANR-11-LABX-0010), DGTM Guyane, Office de l'Eau Guyane and SPYGEN. We are indebted to the Guiana National Park (PAG) and to Wayapi people for transport and logistics on the field. We are also indebted to the Wayapi traditional authorities for authorizing access and sampling on their territory. We also thank SPYGEN staff for the laboratory technical support.
Jérôme Murienne and Sébastien Brosse should be considered as co-senior authors.
ABSTRACT
Environmental DNA (eDNA) has been extensively used to facilitate the characterization of both terrestrial and aquatic communities. However, implementing temporal biodiversity monitoring remains a major challenge requiring a better understanding of how seasonal variations in biological and ecological factors influence eDNA detection. Focusing on a French Guiana river, our research investigates the impact of tropical rainfall on ecological patterns of aquatic and terrestrial communities using eDNA samples. We collected water eDNA samples during the dry and rainy seasons to detect fish, mammals, and amphibians. Between seasons, gamma diversity was stable for fish but increased in the rainy season for mammals and amphibians. This shift is explained by the transport of eDNA from terrestrial to aquatic systems through rainfall (eDNA washout) and by the increase of activity during the rainy season for amphibians. In terms of beta diversity, we found species homogenization between sites, affecting both terrestrial and aquatic taxa during the rainy season. This is driven by the increase in water discharge and decrease in water temperature, leading to eDNA drift over longer distances. We recommend sampling eDNA during the dry season for local community inventories and during the rainy season for regional species monitoring, particularly to maximize terrestrial species detection.
1 Introduction
The use of environmental DNA (eDNA) techniques in ecological sciences has exponentially increased during the last decade (Blackman et al. 2024). Many topics can be addressed with eDNA methods, from the detection of invasive or threatened species to the inventory of whole communities for a large range of taxonomic groups and ecosystems (Banerjee et al. 2021; Yao et al. 2022). Such studies have facilitated the detection of hard-to-detect species using classical methods and complemented biodiversity inventories in megadiverse and remote regions (Pont et al. 2018; Cilleros et al. 2019; Brys et al. 2021; Coutant, Richard-Hansen, et al. 2021; Condachou, Cuenot, et al. 2024). eDNA provides rapid characterization of both terrestrial and aquatic communities (Coutant, Cantera, et al. 2021; Fediajevaite et al. 2021; Polanco Fernández et al. 2021) and enables investigation of the spatial structure of biodiversity associated with natural parameters and anthropic disturbances (Cantera, Coutant, et al. 2022; Coutant et al. 2023; Li et al. 2024). However, studies focusing on temporal variations of eDNA are lacking (Mathieu et al. 2020; Blackman et al. 2024) but are of utmost importance to better understand biodiversity changes over time.
Nevertheless, the interpretation of temporal eDNA data is not straightforward because eDNA detection can be influenced by biological factors such as variations in habitat use. For instance, on the Yangtze River, Jia et al. (2020) found lower eDNA abundance in winter due to the overwintering migration of some fish species. Species reproduction can also influence species detection over time by maximizing detection rates during reproduction periods, as shown for fish communities by Hayami et al. (2020). Species detection probabilities could also be affected by environmental conditions that modulate eDNA transport, sedimentation and degradation in the water (Stewart 2019). Seasonal variations in water conditions, such as discharge, temperature or pH, could also contribute to the detectability of species using eDNA (Milhau et al. 2021). This is probably of particular importance in tropical freshwater systems since they display important variations of water parameters between the dry and rainy seasons (Junk et al. 1989). The few studies investigating temporal monitoring using eDNA have already pointed out this potential issue. Interestingly, they provided contradictory results regarding the impact of variations in water parameters on eDNA detectability. For example, Jia et al. (2020) and Liu et al. (2024), linked the fewer species detection to higher eDNA degradation rates in warm waters. Milhau et al. (2021), suggested that eDNA detection increases with water flow due to the transport of eDNA downstream over longer distances, while Curtis et al. (2021), reported a decrease in eDNA concentrations under higher water flow, suggesting a dilution of eDNA. Such dilution effect was also observed in a tropical environment by Sales et al. (2021) who showed a decrease in fish species richness detected using eDNA after a major rain event. Still, Sales et al. (2021) results remain the only case study reporting diversity changes after heavy precipitation in tropical environments, and seasonal comparisons of species detections using eDNA are currently pending.
Most extant eDNA-based studies that have addressed temporal changes have focused on a single taxonomic group, while species detectability could differ through time between aquatic and terrestrial taxa. For instance, Lyet et al. (2021) demonstrated that mammals' eDNA detection probabilities in the aquatic environment were highly influenced by the intensity of rainfall before the sampling event. Evidence of eDNA transport by rainwater in the aquatic system was also provided for terrestrial invertebrates (Macher et al. 2023) and bacterial communities (Yang et al. 2021). Therefore, we hypothesized that eDNA can be transported from land to rivers through rainwater and refer to this process as “eDNA washout”.
Here, we investigate the effect of seasonality on ecological patterns obtained from eDNA samples. We explored this by collecting eDNA from the Oyapock River (Northeastern Amazonia), which is characterized by an equatorial climate with a typical alternance of dry and rainy seasons. This area also benefits from previous studies on the use of eDNA to detect vertebrate communities from aquatic samples (Cilleros et al. 2019; Coutant, Cantera, et al. 2021; Coutant, Richard-Hansen, et al. 2021; Cantera, Coutant, et al. 2022). In this study, we investigated the effect of seasonality on the measure of fish, mammal, and amphibian diversity patterns (alpha, beta and gamma) using eDNA metabarcoding by sampling the same 20 sites in the dry and the rainy seasons. At the regional scale (gamma diversity), we expect that the seasonal effect should impact terrestrial organisms because higher rainfall during the wet season should wash out terrestrial eDNA into the aquatic environment (Lyet et al. 2021). This should increase gamma diversity for terrestrial species, with a particular increase in the detection of low-water affinity species compared to species more dependent on the aquatic system (living in or close to water, see methods) (Sasso et al. 2017). On the contrary, the gamma diversity of high-water affinity species should be similar for both seasons. At the local scale, we hypothesize that during the rainy season, higher discharge (increasing eDNA transport) combined with lower temperature (decreasing eDNA degradation) should increase species detection distances. This is in line with the findings of Deiner et al. (2016), who demonstrated that in contexts of high-water transport, eDNA samples can integrate information over large spatial scales. We thus expect a higher alpha diversity per site during the rainy season due to the decrease in eDNA degradation, but a lower species dissimilarity (lower beta diversity) between sites, due to the increased downstream transport of eDNA that should homogenize distant communities. We expect all taxonomic groups (fish, mammals and amphibians) to be affected by the increase in detection distances, irrespective of their ecology and water affinity.
2 Materials and Methods
2.1 Study Sites
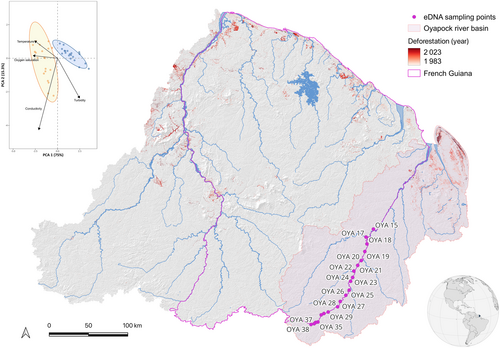
Amazonia is a highly diversified region, within which French Guiana hosts about 200 mammals, 130 amphibians, and 350 fish species (UICN France 2017), including about 200 fish species recorded in the Oyapock River alone (Le Bail et al. 2012). Aquatic environmental samples were collected in twenty sites of the upper Oyapock River (Figure 1). Of the twenty sites sampled, fifteen were located on the main river channel, while the remaining five included one major tributary (OYA17, Camopi River) and four smaller tributaries (OYA15, OYA22, OYA26, and OYA31). The Oyapock is a ca. 400 km long river, and its watershed covers a surface of 26,800 km2 flowing over Amapá state (Brazil) and French Guiana. Samples were collected on the same sites in the dry season (November 2018) and in the rainy season (March 2022) within the Amazonian French Guiana National Park (PAG). This region has a very low human density (< 1 habitant/km2), no cities or agricultural exploitations, and a forest cover of more than 95% (World Resource Institute 2014). As highlighted by the Tropical Moist forest dataset (Vancutsem et al. 2021), this area has not suffered any major disturbance in the past 40 years, with deforestation only occurring in the coastal areas of the region (Figure 1). Therefore, no other change in species communities than seasonal variations is expected between the two sampling years. The study area is moreover of particular interest for evaluating the impact of season on biodiversity patterns because French Guiana harbors a typical equatorial climate with a dry season (from July to November) and a rainy season (from December to June) characterized by strong differences in rainfall. Indeed, at the French Guiana scale, our two sampling events were characterized by different cumulative rainfall levels (November 2018: 210.69 ± 76.10 mm; March 2022: 595.62 ± 237.24 mm) (Meteo France) (Figure S1). To assess the changes in physicochemical water parameters, at each site and sampling event (dry and rainy seasons), water temperature, pH, conductivity, and O2 saturation were recorded with a multiparameter sensor (3620 WTW) as well as water turbidity (TN100, Eutech instruments).
2.2 eDNA Sampling and Extraction
At each of the twenty sites and per season, two eDNA sample replicates were collected by filtering water for 30 min, following recommendations from Cantera et al. (2019). A 500 cm2 polyethersulfone filter with a pore size of 0.45 μm (VigiDNA 0.45 SPYGEN) was used. A peristaltic pump (Vampire sampler, Bürkle) and disposable sterile tubing were used to pump the water through the encapsulated filtering cartridges. The input part of the tube was held a few centimeters below the surface in zones with high water flow. The operator remained downstream from the filtration area to avoid DNA contamination among sites. At the end of the filtration, the cartridge was emptied of water and filled with 80 mL of CL1 preservation buffer (SPYGEN). Cartridges were then stored in the dark and at room temperature until further lab processing. Then, DNA extraction was performed following the procedure described by Pont et al. (2018) and reported in the Supporting Information. Shortly, each filtration capsule was shaken for 15 min at 800 rpm and then centrifuged for 15 min at 15,000xg. Ethanol (33 mL) and sodium acetate (1.5 mL) were added, and samples were stored overnight at −20°C. After a centrifugation step, ATL buffer was added, and samples were incubated for 2 h at 56°C with Proteinase K. Then, DNA extraction was performed following the NucleoSpin Soil kit (MACHEREY-NAGEL GmbH & Co., Düren Germany) starting from step six. The elution was performed by adding 100 μL of SE buffer twice. After DNA extraction, samples were tested for inhibition by qPCR following the protocol in Biggs et al. (2015). Briefly, qPCR was performed in duplicate for each sample. If at least one of the replicates showed a different cycle threshold than expected (at least two cycle thresholds), the sample was considered inhibited and diluted fivefold before the amplification.
2.3 Multi-Species Detection by Metabarcoding
We used the 12S-V5 vertebrates' primers [Forward 5′-TAGAACAGGCTCCTCTAG-3′, Reverse 5′-TTAGATACCCCACTATGC-3′] (Riaz et al. 2011), which have already provided relevant results to describe biodiversity of French Guiana (Kocher, de Thoisy, Catzeflis, Huguin, et al. 2017; Kocher, de Thoisy, Catzeflis, Valière, et al. 2017; Cantera et al. 2019; Coutant, Richard-Hansen, et al. 2021; Cantera, Coutant, et al. 2022; Kocher et al. 2023). The eDNA amplification (12 PCR replicates per sample), high-throughput sequencing and bioinformatics were performed following the procedure described by Coutant, Richard-Hansen, et al. (2021) (see Supporting Information for details). Taxonomic assignment of the MOTUs was performed using the updated local fish reference database of Cilleros et al. (2019) providing reference data for 370 out of the 415 freshwater fish species known in French Guiana. Concerning mammals, we used the same reference database as the one used in Kocher, de Thoisy, Catzeflis, Huguin, et al. 2017, which covers 83% of the mammal species recorded in French Guiana, representing 92% of genera and 100% of families. Finally, the amphibian reference database included 123 species, representing more than 95% of all known amphibian species. MOTUs showing less than 98% similarity to the reference database and with a read count below 50 were removed. When the taxonomic assignment was ambiguous between two species, the MOTU was assigned to the species known to occur in the studied area. If no information about the species distribution was available, sequences were discarded, thus keeping for the ecological analyses only MOTUs assigned to the species level.
2.4 Data Analysis
First, to check the validity of our sampling design, we looked at how similarity in species composition between site pairs declined with increasing geographic distance. Presence/absence matrices of the species detections in each site, including all taxonomic groups, were used to build a species compositional distance matrix between pairs of sites for each season with the vegdist function (vegan 2.6–6.1 R package). Pairwise compositional distances between sites were calculated using the Jaccard index. We used linear regressions to model the relationship between species similarity distances and watercourse distances between sites (in km) for each season. Alpha diversity patterns were investigated by comparing the species richness across sites depending on the season and taxonomic groups. Mammal species were classified into three groups depending on their habitats: arboreal, terrestrial, and semi-aquatic (Coutant, Richard-Hansen, et al. 2021) and thus indirectly their water affinity from low (arboreal species) to high (semi-aquatic species) (Table S1). We classified as “arboreal” the species mainly living in trees based on the study of Coutant, Richard-Hansen, et al. (2021) and specific literature searches on species ecology. Flying species belonging to Chiroptera were considered as arboreal species. Amphibians were classified according to their water affinity, considering the species with no or a single aquatic life stage (egg, tadpole or adult) as having low water affinity. In contrast, the species with two or three aquatic life stages were considered to have a high-water affinity (Table S2).
Species occurrences were calculated for each season as the number of detections across the 40 samples (two samples per site). Gamma diversity was investigated for the three taxonomic groups and considered as the total number of species detected across the study area. To evaluate the efficiency of the sampling effort according to the season, accumulation curves were generated using the Chao2 method (Chao et al. 2014) (vegan 2.6–6.1 R package) (Figure S2).
Beta diversity was investigated by generating a distance matrix based on the Jaccard index for all pairs of sites using the “vegdist” function (vegan 2.6–6.1 R package). Principal coordinates analysis (PCoA) based on the Jaccard dissimilarity index was then performed for all the taxonomic groups using the “cmdscale” function (stats 3.6.2 package). To investigate the effect of seasonality on species community composition, a Permutational Multivariate Analysis of Variance (PERMANOVA) applying the Jaccard dissimilarity index was performed with the function “adonis2” (vegan 2.6–6.1 R package). Dispersion between samples of the same season was investigated with the BETADISPER function (vegan 2.6–6.1 R package). The “betapart” package (version 1.6) was used to calculate beta diversity and to partition it into nestedness and turnover components (Baselga and Orme 2012).
3 Results
For the 80 samples analyzed in this study, a total of 55,575,063 reads were obtained after the sequencing step. After bioinformatics processing, 41,487,505 reads were retained, corresponding to species belonging to the classes Actinopterygii, Chondrichthyes, Amphibia, and Mammalia.
3.1 Characterization of Studied Sites
Results for the distance decay showed that similarity significantly decreased with watercourse distance between site pairs in the dry season (R2 = 0.32, p < 0.001) and the rainy season (R2 = 0.23, p < 0.001) (Figure S3). Physico-chemical water parameters and regional meteorological conditions differed between the two seasons (Figure 1, Table S3). During the dry season, the sampled sites were characterized by a mean water temperature of 27.8°C (sd:±1.6°C), pH 7.3 (±0.2), 95% of O2 saturation (±4.3%) and a turbidity of 7.8 NTU (±3.9 NTU). During the rainy season, the mean water temperature (24.5°C ± 0.3°C), pH (6.3 ± 0.4) and O2 saturation (82.7% ± 5.8%) were lower. The mean turbidity across the sites was higher in the rainy season (15.8 ± 5.3 NTU).
3.2 Variations of Gamma and Alpha Diversity Across Seasons
The gamma diversity (number of detected species across all sample sites) was higher during the rainy season for mammals and amphibians, with 87 mammals and 42 amphibians detected during the rainy season, while 53 mammals and 17 amphibians were found during the dry season (see detailed species list in Tables S1, S2 and S4). Fish gamma diversity remains constant between seasons, with 117 species detected during the rainy season and 114 during the dry season.
Considering alpha diversity, during the dry season 75 ± 12 fish, 9 ± 4 mammals, and 4 ± 1 amphibian species were detected on average by site. During the rainy season 90 ± 14 fish, 23 ± 8 mammals, and 15 ± 4 amphibian species were detected per site. For the three taxa, more species were thus detected per site during the rainy season (t-test, p < 0.001, for all three groups) (Figure 2).

Accumulation curves showed similar Chao's asymptomatic species richness between seasons for the three groups, with a value of 120.09 ± 7.37 fish species in the dry season and 118.46 ± 2.24 fish species in the rainy season (Figure S2). For mammals, the Chao's asymptomatic species richness was 70.61 ± 11.37 in the dry season versus 75.08 ± 2.13 in the rainy season. For amphibians, the asymptomatic species richness was 52.1 ± 24.97 in the dry season versus 49.96 ± 8.08 in the rainy season.
In the rainy season, 66% of the fish taxa were detected in more than 30 out of the 40 samples versus 45% during the dry season. The most common and rare species were the same during both seasons (Figure S4). The fish species detected across seasons were highly conserved (93% of common species) (Figure 2). Only 1% of the species were solely detected during the dry season (Anablepsoides gaucheri) and 3% of the species were only detected during the rainy season (Moenkhausia grandisquamis, Gasteropelecus sternicla, Phenacogaster wayampi, and Hypopygus lepturus). Concerning mammals, the frequencies of occurrences were lower during the dry season except for the South American tapir (Tapirus terrestris), which was detected in all sites regardless of the season (Figure S4).
The percentage of species in common between seasons was lower than for fish (58% vs. 93%), with 37% of species only detected during the rainy season (Figure 2). Differences in mammal occurrences through seasons were variable depending on their habitats with significantly more arboreal mammals detected during the rainy season than during the dry season (Wilcoxon test, p < 0.01) (Figure 3). Concerning the semi-aquatic and terrestrial mammals, no significant differences were observed in the species occurrence depending on seasons. The most important change in the number of species detected across seasons (N = 10 during the rainy season vs. N = 3 during the dry season) was observed for the bats belonging to the Phyllostomidae family.
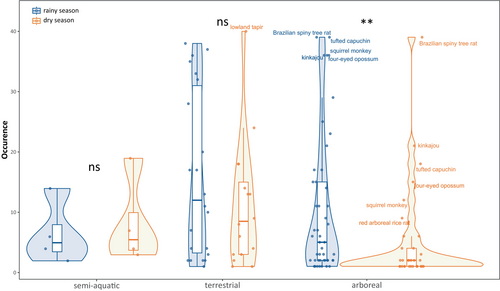
The vast majority (54%) of the detected amphibian species are linked to the aquatic environment at least during more than one life stage (egg, tadpole, adult). Species with terrestrial modes of development (endotrophic i.e., without free-feeding larvae and/or nidicolous i.e., remaining in the nest until metamorphosis, phytotelmous i.e., larvae developing in small water collections in epiphytic plants or trunks…) are almost absent. The few detected endotrophic species are aquatic e.g., Pipa spp., or associated with damp habitats near streams like Caecilia gracilis. During the rainy season, more low-water affinity species are detected than during the dry season (Wilcoxon test, p < 0.05) (Figure 4). Pipa pipa still presents a high detection rate during the dry season because of the aquatic affinity of the adult life stage. During the dry season, only two species with strong adult aquatic affinity (Lithobates palmipes and Rhinella marina) are found in more than 30 samples out of the 40, versus eight species in the rainy season (Figure S4). Only 38% of amphibian species are found in both seasons, whereas 61% of the species were only detected in the rainy season (Figure 2).

3.3 Variations of Beta Diversity Across Seasons
For all taxonomic groups, beta diversity between samples was significantly higher during the dry season than during the rainy season (Wilcoxon test, p < 0.001) (Table 1). For fish and mammals, in the dry season, species composition was mainly explained by the species turnover between sites (turnover component of beta diversity), while the difference in species richness between sites (nestedness component of beta diversity) remained low. During the rainy season, the turnover component was significantly lower (Wilcoxon test, p < 0.001). This reveals that during the rainy season, fish and mammal species composition was more homogeneous between sites (low beta diversity) due to a lower species turnover between sites. Changes in beta diversity for amphibians were explained by the significant increase of the nestedness component of biodiversity during the dry season (Wilcoxon test, p < 0.01), thus indicating a prominent effect of species richness differences between sites in the amphibians' beta-diversity patterns.
| Dry season | Rainy season | Wilcoxon test p | |
|---|---|---|---|
| Fish | |||
| Beta-diversity | 0.27 ± 0.09 | 0.17 ± 0.09 | p < 0.001 |
| Turnover | 0.21 ± 0.09 | 0.07 ± 0.06 | p < 0.001 |
| Nestedness | 0.05 ± 0.04 | 0.09 ± 0.08 | p < 0.001 |
| Mammals | |||
| Beta-diversity | 0.64 ± 0.17 | 0.47 ± 0.14 | p < 0.001 |
| Turnover | 0.49 ± 0.21 | 0.33 ± 0.14 | p < 0.001 |
| Nestedness | 0.14 ± 0.14 | 0.15 ± 0.13 | p > 0.05 |
| Amphibians | |||
| Beta-diversity | 0.52 ± 0.25 | 0.46 ± 0.13 | p < 0.001 |
| Turnover | 0.35 ± 0.32 | 0.36 ± 0.15 | p < 0.01 |
| Nestedness | 0.16 ± 0.17 | 0.09 ± 0.08 | p > 0.05 |
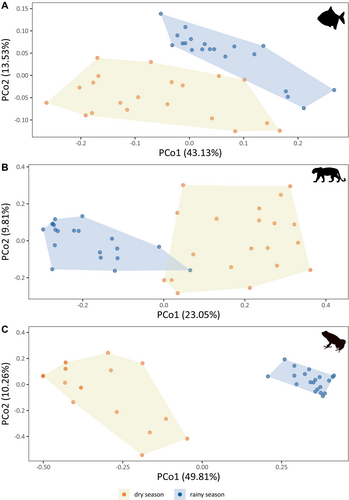
The amount of variation captured by the first two axes of each PCoA was above 30% for the three taxonomic groups, thus providing a good two-dimensional representation of the sample pairs according to their community composition (Figure 5). For all taxonomic groups (fish, mammals and amphibians), the season has a significant effect on species composition (PERMANOVA(fish), F = 8.89, p < 0.001; PERMANOVA(mammal), F = 8.47, p < 0.001; PERMANOVA(amphibian), F = 32.60, p < 0.001). For fish and mammals, the sites dispersion was significantly higher during the dry season (BETADISPER(fish), F = 15.13, p < 0.001; BETADISPER(mammal), F = 30.64, p < 0.001) but not for amphibians (BETADISPER(amphibian), F = 0.66, p > 0.05). In contrast, the sites were clustered together in the rainy season, suggesting a homogenization of species composition during this period.
4 Discussion
Investigating the seasonal variations of ecological patterns inferred using eDNA revealed contrasting biodiversity patterns depending on the species' water affinity and the considered biodiversity scale (regional or local).
At the regional scale (gamma diversity), the inferred diversity of fish was similar for both seasons (117 species detected during the rainy season and 114 during the dry season), reinforcing previous results about the robustness and replicability of the eDNA method for assessing French Guiana fish biodiversity (Cantera et al. 2019; Cantera, Decotte, et al. 2022; Coutant, Cantera, et al. 2021). This result showed no evidence of long-distance seasonal migration, thus contrasting with the Amazon River fish fauna, where, for instance, large-sized Pimelodidae (Brachyplatystoma spp.) and Curimatidae (Prochilodus spp.) are known to migrate over hundreds of kilometers (Duponchelle et al. 2021). This is not the case in the studied area, where Curimatidae are captured throughout the year by local people, and Brachyplatystoma catfishes are only known from the lower estuarine region that was not investigated in this study. In addition, amphidromous species (i.e., species migrating between sea and freshwater) do not occur in French Guiana, and long-distance migrations have never been observed. Information about species migratory habits is still incomplete in the rivers of Northern Amazonia (including French Guiana), and continued temporal eDNA monitoring may help fill this gap. Nevertheless, detecting fish migration using eDNA can be complicated as migration rarely involves entire populations but rather certain life stages (reproductive individuals or juveniles). Migration patterns could be detected only with reliable quantitative data, which we currently lack in natural environments (Yates et al. 2019).
For mammals and amphibians, the number of species detected at the regional scale peaked during the rainy season. For amphibians, this trend showed an even more marked difference between seasons, probably due to behavioral and ecological mechanisms specific to this taxonomic group. For instance, during the dry season, most amphibian species reduce their activity and movement, and only very few species are reproductively active (Fouquet et al. 2024). The vast majority of amphibian species reproduce during the rainy season and many breed in ponds and puddles, often nearby rivers, but very few species breed directly in the rivers. These water bodies probably concentrate eDNA since they are used by breeding adults releasing gametes and then by their tadpoles. After rain events, eDNA can thus be washed out from the fluvial annexes into the river, increasing species detectability. Conversely, some Amazonian species with terrestrial development are not detected in either seasons. This corresponds to species with particular life stages that do not rely on water systems, such as species with endotrophic larvae development or species laying eggs in plants. Sasso et al. (2017), found similar results in Brazilian streams, with no eDNA detection of terrestrial species and high detection rates for species with high dependence on aquatic habitats. On the contrary, mammal species are still active during the dry season and mainly rely on rivers for their water sources. In addition, mammals are not known to change their spatial occupancy between seasons (Emmons and Feer 1997), and in water systems, mammal eDNA is rare during the dry season, reducing species detectability (Coutant, Richard-Hansen, et al. 2021). Thus, there is a clear increase in mammal eDNA washout (i.e., the transport of terrestrial eDNA to the aquatic ecosystem through rainfall) in rivers during the rainy season, increasing mammal detectability.
Overall, the difference in mammal and amphibian gamma diversity between seasons can be explained by the eDNA washout, although seasonal variations in species behavior can also affect eDNA availability. Indeed, differences in local (alpha) diversity between seasons for mammals and amphibians were mainly due to the increase in low-water-affinity species detection during the rainy season. Arboreal mammal species presented higher detection rates during the rainy season as well as amphibian species with none or a single aquatic life stage. This reinforces the eDNA washout hypothesis, highlighting that DNA coming from the terrestrial environment is retrieved in the river during the rainy season. Such results have a particular significance for terrestrial fauna inventories using aquatic eDNA. Variations in eDNA detection according to species diet or behavior (Sales et al. 2019; Lyet et al. 2021; Mena et al. 2021) refer to samples collected during the dry season. Such limitations might be lifted, or at least reduced, by sampling terrestrial species during the rainy season. Moreover, further studies should investigate the spatial and temporal scale of the eDNA signal for terrestrial species to assess the washout distance between terrestrial and aquatic environments, as well as eDNA persistence in the terrestrial environment, which could be much longer than in the water. For instance, Leempoel et al. (2020) showed that mammal eDNA can be stored in the soil for 30 to 150 days, thus providing a longer-term view of the terrestrial fauna than that of aquatic species.
Biodiversity differences between sites (beta-diversity) peaked during the dry season. Such species replacement between sites is consistent with previous fish studies (Cilleros et al. 2016). This is also consistent with Cantera, Decotte, et al. (2022), demonstrating that during the dry season, the eDNA signal provides a spatially limited inventory of the fish fauna. Condachou, Coutant, et al. (2024) confirmed this by demonstrating a short downstream detection distance (less than 100 m) for an Amazonian stream armored catfish (Harttiella lucifer). In contrast, during the rainy season, species assemblages appear more similar between sites (with a decline of beta diversity), demonstrating a homogenization between sites in fish, amphibians, and mammals. Such a trend is probably driven by the drastic increase in water discharge during the rainy season, which could lead to eDNA drift over longer downstream distances (Wood et al. 2021; Van Driessche et al. 2023; Condachou, Coutant, et al. 2024; Pont 2024). Moreover, the lower temperature and solar radiation during the rainy season also probably contribute to slowing the eDNA degradation rate (Lamb et al. 2022; Liu et al. 2024), thus reinforcing the homogenization of the communities detected using eDNA. The seasonal reproduction of fish and amphibians with the release and drift of gametes, eggs, and larvae could also reinforce species detection (Milhau et al. 2021). However, since the homogenization signal is consistent for both fish, amphibians, and mammals, it can reasonably be considered that longer downstream eDNA drift is the primary factor explaining beta diversity differences between seasons.
Those results have a particular significance for biodiversity management and conservation. While dry-season eDNA samples should be preferred in studies focusing on local community inventories, sampling eDNA during the rainy season is more appropriate for regional monitoring of biodiversity by providing spatially extended biodiversity information. Sampling eDNA during the rainy season also maximizes species detection for low water affinity species through eDNA washout and should be preferred when searching for rare and/or exclusively terrestrial mammals and amphibians (e.g., endotrophic/direct-developing or phytotelmous frogs). The interpretation of these results should account for the possibility of natural biodiversity changes across years. Indeed, there is a two-year interval between the two sampling events. Despite this gap, we remain confident in our findings as the study region has a very low human density and has not suffered any major deforestation, which likely minimizes significant biodiversity shifts over a short time frame.
This study reports seasonal differences in species detectability. This raised the importance of integrating detection probabilities in eDNA studies. Indeed, season is not the only potential factor that could induce changes in species detectability. Species inventory provided by eDNA is prone to both false negatives (where a species is present but not detected) and false positives (where a species is detected but not present) (Lahoz-Monfort et al. 2016; Burian et al. 2021). False positives resulting from contamination, failures of primer specificity, or technical issues are now well-recognized and can be managed with appropriate laboratory and bioinformatics workflows (Tingley et al. 2021). Nevertheless, it is important to note that some detections labeled as false positives may reflect biological realities, such as the transport of eDNA, rather than technical mistakes (Darling et al. 2021). Statistical approaches, such as occupancy modeling, can help address these challenges by estimating detection probabilities (Nichols et al. 2008; Dorazio and Erickson 2018; Peixoto et al. 2023; Donovan et al. 2024). Hydrologic-based models have also recently been developed to consider eDNA transport and degradation for estimating eDNA detection probabilities (Carraro and Altermatt 2024; Pont 2024). Especially in temporal and long-term studies, estimating detection probabilities using an appropriate sampling design is crucial to increase the robustness of the results. Despite those limitations, eDNA metabarcoding remains a fast and effective method to inventory both aquatic and terrestrial biodiversity, and our results offer a way to consider local (during dry periods) and regional (during wet periods) faunistic inventories using eDNA.
Author Contributions
C.C., J.M., and S.B. conceived the ideas and designed the methodology; J.M., S.B., and O.C. collected the data; A.V. conducted the laboratory work; C.C., J.M., S.B., and A.F. analyzed the data. All authors contributed critically to the drafts and gave final approval for publication.
Conflicts of Interest
Alice Valentini is a research scientist in a private company specialized in the use of eDNA for biodiversity monitoring (SPYGEN).
Open Research
Data Availability Statement
Supplementary materials including Supporting Information about the materials and methods, the mammals, amphibians, and fish species list (Tables S1, S2 and S4 respectively) and a summary of water physico-parameters and coordinates of sampled sites (Table S3) underlined the main results of the study are provided in the online version of the article. Raw species data detection for each site is provided in the OTU_table.xls Supporting Information. The Illumina raw sequence data for the 2018 (dry season) and 2022 (rainy season) are available on Figshare (https://doi.org/10.6084/m9.figshare.13739086.v7; https://doi.org/10.6084/m9.figshare.27232593, respectively).




