Journal list menu
Export Citations
Download PDFs
Table of Contents
Can single cell RNA sequencing reshape the clinical biochemistry of hematology: New clusters of circulating blood cells
- First Published: 12 December 2021
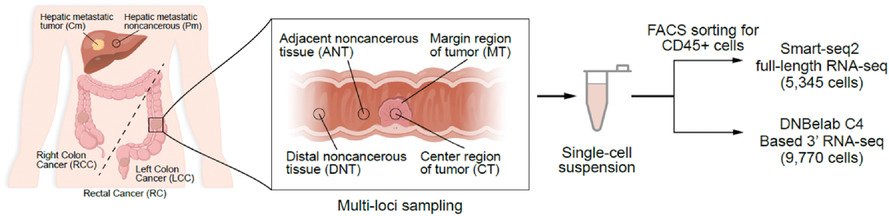
Multiregion single-cell sequencing reveals the transcriptional landscape of the immune microenvironment of colorectal cancer
- First Published: 01 January 2021
Single-cell analyses reveal suppressive tumor microenvironment of human colorectal cancer
- First Published: 06 June 2021
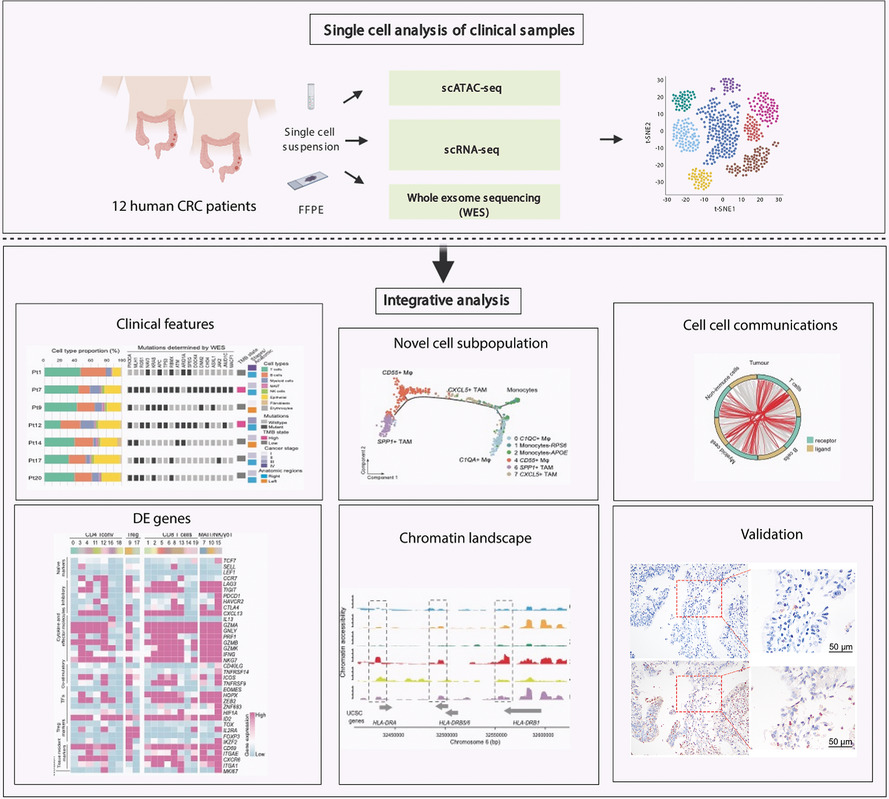
1) Single-cell profiling of clinical samples of 12 human colorectal cancer patients.
2) Tumor mutational burden states of human CRC contribute to distinct immune profile patterns.
3) Single-cell analysis in human CRC identified phenotypic and functional diversity of tumor-associated macrophages and T cells.
Diversity and intratumoral heterogeneity in human gallbladder cancer progression revealed by single-cell RNA sequencing
- First Published: 27 June 2021
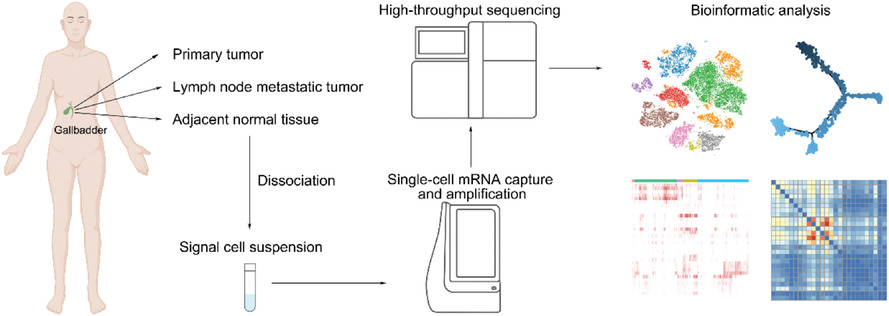
-
Abundant immune cells in adenocarcinoma and squamous cell carcinoma while stromal cells in neuroendocrine neoplasms were enriched.
-
Immunosuppressive microenvironment characterized as exhausted T cells and APOE+ macrophages.
-
Endothelial cells (KDR+ and ACKR1+) indicated that angiogenesis and lymphangiogenesis were involved in GC.
-
There were remarkable ligand–receptor interactions between endothelial, primary GC cells, and macrophages.
Landscape and dynamics of single tumor and immune cells in early and advanced-stage lung adenocarcinoma
- First Published: 09 March 2021
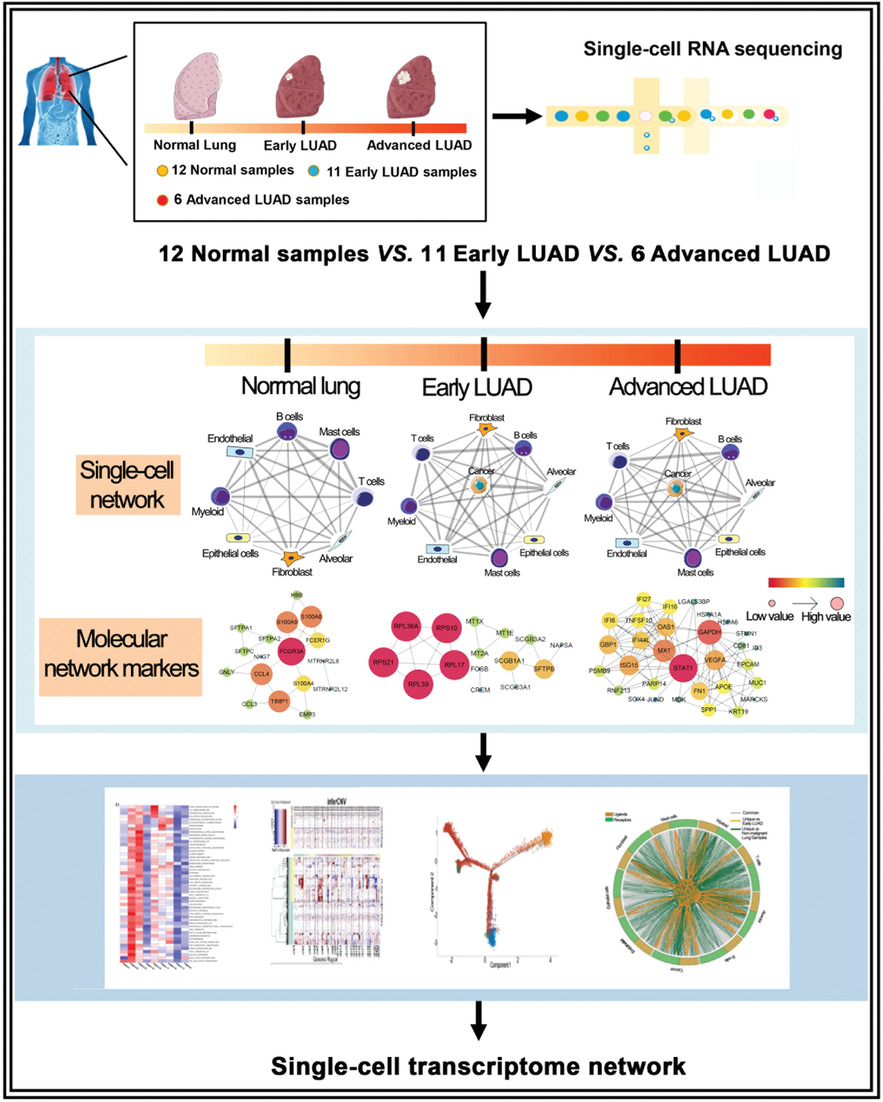
- Our study comprehensively studied the tumor microenvironment (TME)and intra-tumor heterogeneity (ITH) in different stages of LUAD by both scRNA-seq and bulk RNA-seq analyses.
- Based on these high-quality cells derived from different tissues, our results revealed the cellular diversity and molecular complexity of cell lineages in different stages of LUAD.
- This study, which serves as a basic framework and valuable resource, can facilitate exploration of the pathogenesis of LUAD and identify novel therapeutic targets in the future.
Combining single-cell sequencing to identify key immune genes and construct the prognostic evaluation model for colon cancer patients
- First Published: 19 July 2021
Neoadjuvant chemotherapy alters peripheral and tumour-infiltrating immune cells in breast cancer revealed by single-cell RNA sequencing
- First Published: 17 December 2021
A cellular census of human peripheral immune cells identifies novel cell states in lung diseases
- First Published: 24 November 2021
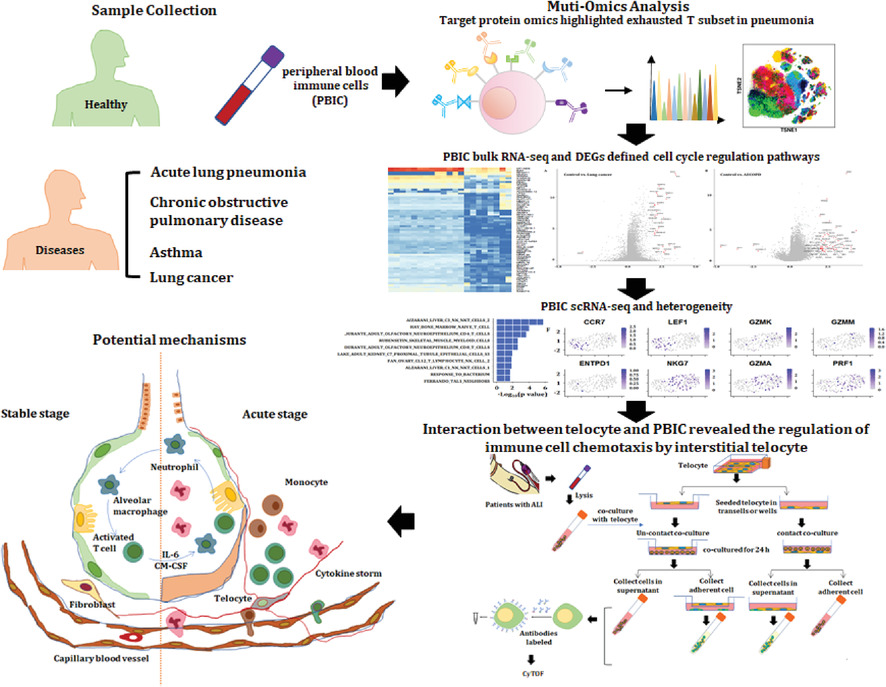
1. New clusters and functions of circulating immune cells can be an important source to understand molecular mechanisms, identify disease-specific biomarkers, and develop new target therapies.
2. The difference of biological processes and pathways of PBICs by single-cell sequencing and RNAseq varies among diseases.
3. The interstitial cells could alter the differentiation and development of the immune cell types and immune function.
Single-cell RNA sequencing infers the role of malignant cells in drug-resistant multiple myeloma
- First Published: 17 December 2021
Integrated single-cell RNA sequencing analyses suggest developmental paths of cancer-associated fibroblasts with gene expression dynamics
- First Published: 19 July 2021