Journal list menu
Export Citations
Download PDFs
Cover Image, Volume 39, Issue 11
- Page: i
- First Published: 11 October 2018
On the Cover: This cover image is based on the Special Article Unique aspects of sequence variant interpretation for inborn errors of metabolism (IEM): The ClinGen IEM Working Group and the Phenylalanine Hydroxylase Gene by Diane B. Zastrow et al., Pages 1569–1580. DOI: 10.1002/humu.23649.
Issue Information
- Pages: 1469-1472
- First Published: 11 October 2018
ClinGen and ClinVar – Enabling Genomics in Precision Medicine
- Pages: 1473-1475
- First Published: 11 October 2018
The ClinGen Epilepsy Gene Curation Expert Panel—Bridging the divide between clinical domain knowledge and formal gene curation criteria
- Pages: 1476-1484
- First Published: 11 October 2018
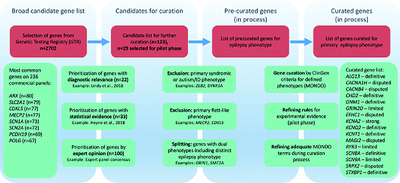
Epilepsy is emerging as a frequent indication for diagnostic genetic testing. The ClinGen Epilepsy Gene Curation Expert Panel is tasked with connecting the domain of traditional clinical epileptology and the rapidly evolving area of diagnostic genetic testing. We identify precise phenotype definitions and phenotypic boundaries, gene selection strategies, and rules functional models for seizure disorders as critical components of the gene curation process. We demonstrate that curation for epilepsy-associated genes is feasible and suggest epilepsy-specific conventions.
Assessing the gene–disease association of 19 genes with the RASopathies using the ClinGen gene curation framework
- Pages: 1485-1493
- First Published: 11 October 2018
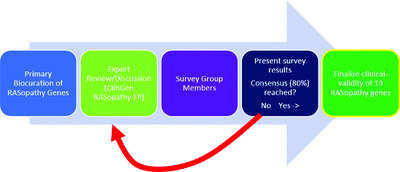
The ClinGen RASopathy Expert Panel assessed the published and other publicly available evidence supporting the association of 19 genes with RASopathy conditions using ClinGen's semiquantitative literature curation method. Our group assessed the evidence implicating each gene's association with Noonan syndrome, Costello syndrome, cardiofaciocutaneous syndrome, Noonan syndrome with multiple lentigines, and Noonan-like syndrome with loose anagen hair. Our results will guide clinicians, patients and researchers in their understanding of the gene-disease relationships within the RASopathies.
The progression of the ClinGen gene clinical validity classification over time
- Pages: 1494-1504
- First Published: 11 October 2018
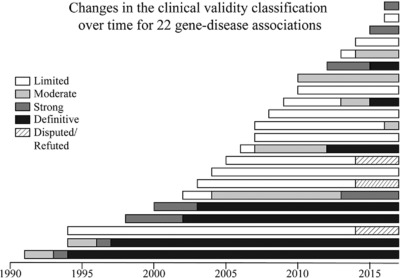
Maintaining the most current ClinGen gene-disease clinical validity classifications requires an appropriate timeline for re-evaluating curated gene-disease associations. In order to provide guidance on how often a gene-disease association should be re-evaluated, a retrospective analysis of 30 gene curations was performed at one-year intervals to identify trends in the classification over time and factors that influenced changes in classification. This study provides key consideration points for determining the most effective timeline for re-evaluating gene-disease associations.
On the verge of diagnosis: Detection, reporting, and investigation of de novo variants in novel genes identified by clinical sequencing
- Pages: 1505-1516
- First Published: 11 October 2018
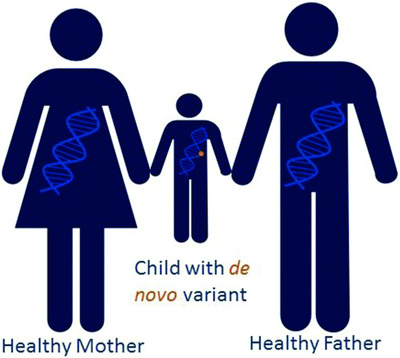
In this study, we investigate the detection, reporting and re-interpretation of de novo variants in novel genes uncovered by clinical next-generation sequencing. These novel genes were subjected to ClinGen scoring to assess the strength of gene-disease relationships and facilitate accurate molecular diagnosis.
Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion
- Pages: 1517-1524
- First Published: 07 September 2018
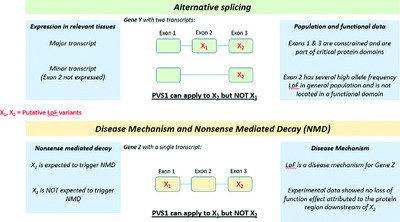
We provide guidance for PVS1 usage that takes into consideration all aspects of putative loss of function (LoF) variants, including type, location, and annotation, and the disease mechanism of the genes they affect. We demonstrate how the combination of these variant and gene attributes can lead to varied PVS1 strength levels. Finally, we evaluate the refined criterion using > 50 LoF variants in several genes and diseases.
Updated recommendation for the benign stand-alone ACMG/AMP criterion
- Pages: 1525-1530
- First Published: 11 October 2018
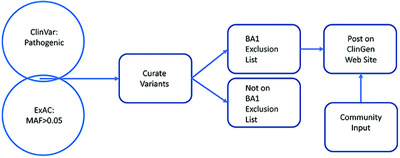
The ACMG/AMP variant pathogenicity recommendations include a stand-alone rule BA1 (variant with MAF >0.05 is benign). We cross-referenced ClinVar and ExAC for variants with MAF >0.05 and reasonable evidence for pathogenicity, identifying nine (four pathogenic and five VOUS) variants as a part of an initial exclusion list. We specify how individuals can amend this list. Additionally, we clarified terms, criteria and assumptions about rule BA1 and specify the criteria that experts should use when setting lower gene-specific BA1 thresholds.
Quantifying the potential of functional evidence to reclassify variants of uncertain significance in the categorical and Bayesian interpretation frameworks
- Pages: 1531-1541
- First Published: 10 August 2018
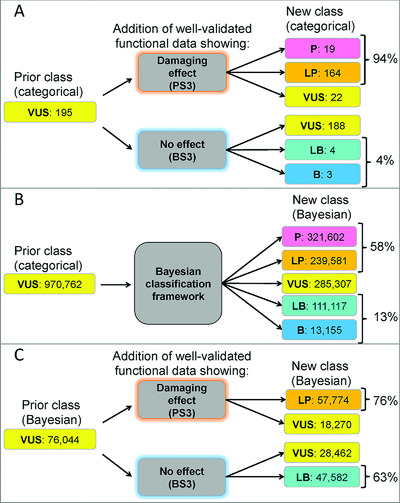
Functional assays have the potential to resolve evidence gaps restricting variants to the category of uncertain significance, but assay development and validation is time consuming and costly. Theoretical algorithmic analysis and an examination of ClinGen Expert Panel variant curations clarify the utility of functional evidence for missense VUS reclassification in the categorical and quantitative clinical variant interpretation frameworks. This analysis lays the groundwork for identifying genes and variants that stand to benefit the most from functional data and thus an evidence-based method for prioritizing assay development and validation.
Integrating somatic variant data and biomarkers for germline variant classification in cancer predisposition genes
- Pages: 1542-1552
- First Published: 11 October 2018
Specifications of the ACMG/AMP variant curation guidelines for the analysis of germline CDH1 sequence variants
- Pages: 1553-1568
- First Published: 11 October 2018
Germline pathogenic variants in the CDH1 gene cause hereditary diffuse gastric cancer and lobular breast cancer. The ClinGen CDH1 Expert Panel developed and implemented rules for CDH1 variant curation, providing the genetic community with a gene-specific framework for the classification of variants identified in this clinically actionable gene. Overall, the Expert Panel specifications resulted in reduced variants of uncertain significance and facilitated resolution of variants with conflicted assertions in ClinVar.
Unique aspects of sequence variant interpretation for inborn errors of metabolism (IEM): The ClinGen IEM Working Group and the Phenylalanine Hydroxylase Gene
- Pages: 1569-1580
- First Published: 11 October 2018
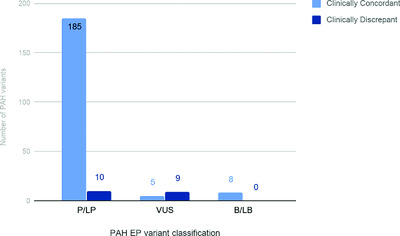
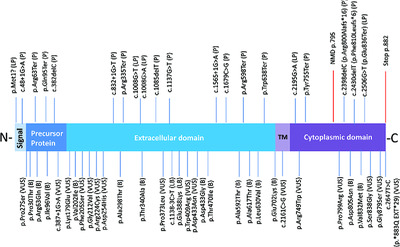
The ClinGen Inborn Errors of Metabolism Working Group chose phenylalanine hydroxylase (PAH) deficiency to pilot metabolic-specific ACMG-AMP variant interpretation guidelines. A PAH variant curation expert panel adjusted existing ACMG-AMP rules based on disease (6) or strength (5) or both (2). Disease adjustments include allele frequency thresholds, functional assay thresholds, and phenotype-specific guidelines. Our validation of PAH-specific variant interpretation guidelines are presented. Using these guidelines 714 PAH variants have been curated and will be submitted to ClinVar.
Gene-specific criteria for PTEN variant curation: Recommendations from the ClinGen PTEN Expert Panel
- Pages: 1581-1592
- First Published: 11 October 2018
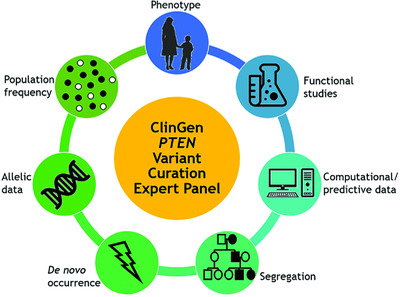
The ClinGen PTEN Expert Panel (EP) includes clinicians, researchers, and molecular diagnosticians with PTEN-related expertise who collaborated to create PTEN-specific variant classification criteria using the framework of the 2015 ACMG/AMP Sequence Variant Interpretation Guidelines. This article describes the development of these criteria and their application for variant classification. These PTEN-specific criteria and the classification assertions made by the EP should prove helpful for laboratories and others who curate PTEN variants.
Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss
- Pages: 1593-1613
- First Published: 11 October 2018

The Hearing Loss Variant Curation Expert panel was created within ClinGen with the goals of standardizing variant interpretation in hearing loss, resolving existing discrepancies in submitted variant classifications in ClinVar and providing expert variant classifications for hearing loss. Here we present specifications of the ACMG/AMP variant interpretation guidelines for hearing loss and the results of a pilot project in which these specifications were applied to 51 variants in the CDH23, COCH, GJB2, KCNQ4, MYO6, MYO7A, SLC26A4, TECTA, and USH2A genes.
ClinGen Variant Curation Expert Panel experiences and standardized processes for disease and gene-level specification of the ACMG/AMP guidelines for sequence variant interpretation
- Pages: 1614-1622
- First Published: 11 October 2018
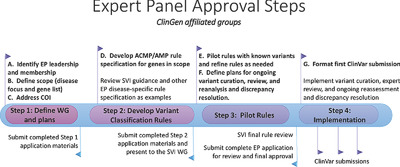
ClinGen is organizing Variant Curation Expert Panels (VCEPs) to develop specifications to the ACMG/AMP guidelines for genes or diseases of interest, interpret variants according to these guidelines, and publish the expert interpretations through the publicly available ClinVar database. A stepwise process was iteratively developed for ClinGen VCEPs to apply to submit variant assertions to ClinVar at the Expert Panel level of review. Other groups that wish to assemble as VCEPs are encouraged, though not required, to follow these steps.
ClinVar at five years: Delivering on the promise
- Pages: 1623-1630
- First Published: 11 October 2018
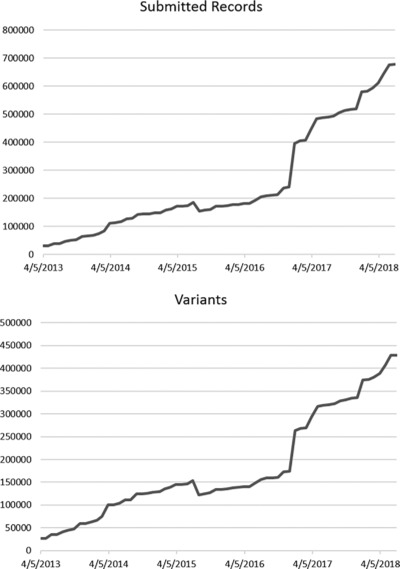
Launched in 2013 at the National Center for Biotechnology Information, National Institutes of Health, ClinVar is a public database for clinical laboratories, researchers, expert panels, and others to share their interpretations of variants with their evidence. The database holds 600,000 submitted records from 1,000 submitters, representing 430,000 unique variants. Building on the platform of providing transparency and leveraging aggregation of variant interpretations, ClinVar is now well positioned to help the clinical genetics community improve interpretations.
ClinVar database of global familial hypercholesterolemia-associated DNA variants
- Pages: 1631-1640
- First Published: 11 October 2018
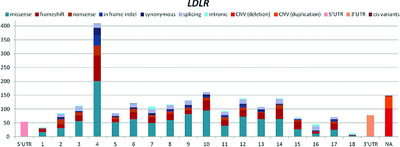
Here, we present the recent efforts made by the Clinical Genome Resource consortium, along with various global familial hypercholesterolemia (FH) researchers, to update the number and characterization of FH-associated variants now present on the ClinVar database. Specifically, we break down the number of FH variants hosted on ClinVar by gene, location, type, and classification; in addition to providing variant-level characterizations. We then discuss the implications learned from these variant-level and aggregate results.
Scaling resolution of variant classification differences in ClinVar between 41 clinical laboratories through an outlier approach
- Pages: 1641-1649
- First Published: 11 October 2018
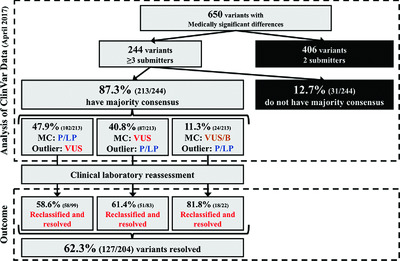
To facilitate resolution of sequence variant classification differences in ClinVar, clinical laboratories were sent a custom report of outlier interpretations and encouraged to reassess variants. Among 650 variants with medically significant differences between laboratories, 31% (204 variants) had an outlier classification, of which 62.3% (127 variants) were reclassified and resolved upon outlier laboratory reassessment alone. This study demonstrates a scalable approach to tackle a portion of classification resolution and capitalizes on the value of data sharing within ClinVar.
Copy number variant discrepancy resolution using the ClinGen dosage sensitivity map results in updated clinical interpretations in ClinVar
- Pages: 1650-1659
- First Published: 10 August 2018
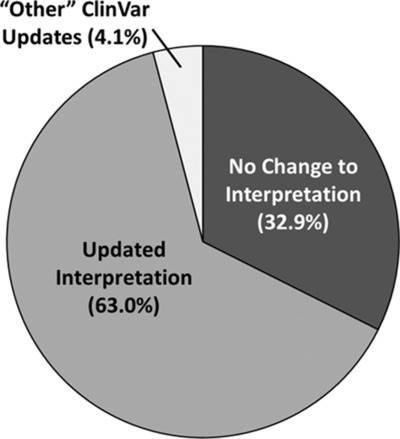
The ClinGen Dosage Sensitivity (DS) Map provides evidence-based assessments of the haploinsufficiency and triplosensitivity of genes/genomic regions. We identified 251 clinical copy number variants (CNVs) in ClinVar that overlapped known DS genes/regions but were not interpreted as “likely pathogenic” or “pathogenic;” these were sent back to their original laboratories for re-evaluation. Of the 246 that were re-evaluated, 63.0% resulted in updated classifications, showing that the ClinGen DS Map can be an effective initial step in CNV classification discrepancy resolution.
The value of genomic variant ClinVar submissions from clinical providers: Beyond the addition of novel variants
- Pages: 1660-1667
- First Published: 11 October 2018
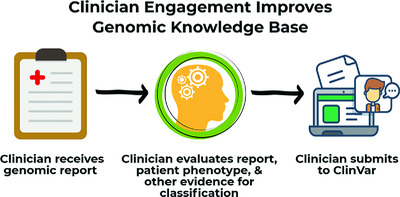
Clinicians can contribute to variant interpretation improvements by submitting variants to ClinVar as “provider interpretations.” Geisinger's Autism & Developmental Medicine Institute has completed a submission of 303 sequence and copy number variants to ClinVar. Clinician engagement in data sharing increases the number of novel variants in ClinVar that have useful clinical descriptions and relevant clinical correlation data and enables the clinician perspective and judgment to be included in variant curation efforts.
ClinGen's GenomeConnect registry enables patient-centered data sharing
- Pages: 1668-1676
- First Published: 11 October 2018
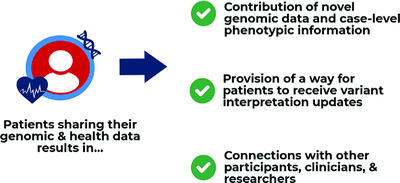
The GenomeConnect patient registry engages patients in data sharing of genomic and phenotypic data. This data sharing allows for the dissemination of novel genomic data, sharing of case-level information, provision of a method to receive variant interpretation updates, and connection of participants with one another and with other stakeholders. By engaging patients, more information is contributed to the public knowledge base benefiting both patients and the genomics community.
Evidence-based assessments of clinical actionability in the context of secondary findings: Updates from ClinGen's Actionability Working Group
- Pages: 1677-1685
- First Published: 11 October 2018
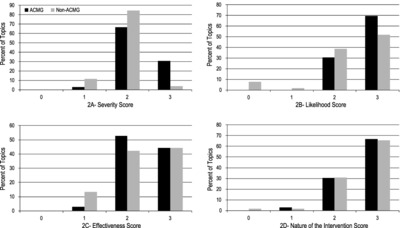
ClinGen's Actionability Working Group (AWG) evaluates four domains of clinical actionability for potential secondary findings: severity and likelihood of the outcome, effectiveness and nature of the intervention. AWG scores were compared between genes/disorders recommended for return as secondary findings by the American College of Medical Genetics and Genomics (ACMG) with those not currently recommended (non-ACMG). Disorders recommended by the ACMG scored higher on outcome-related domains (greater severity and likelihood) but were not different for intervention domains (and effectiveness and nature).
ClinGen advancing genomic data-sharing standards as a GA4GH driver project
- Pages: 1686-1689
- First Published: 11 October 2018

The Clinical Genome Resource (ClinGen) has joined with the Global Alliance for Genomics and Health (GA4GH) to support the development of open, freely-available technical standards and regulatory frameworks for secure and responsible sharing of genomic and health-related data. In its capacity as one of the 15 inaugural GA4GH “Driver Projects,” ClinGen is providing input on the key standards needs of the global genomics community, and has committed to participate on GA4GH Work Streams to help develop and test drive GA4GH deliverables.
ClinGen Allele Registry links information about genetic variants
- Pages: 1690-1701
- First Published: 11 October 2018
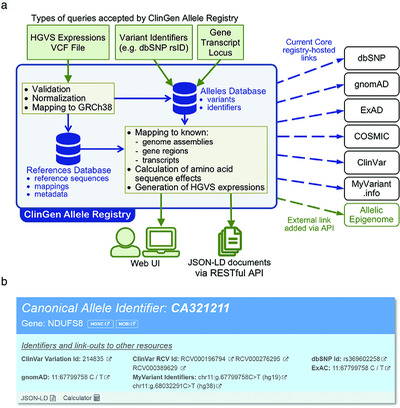
The ClinGen Allele Registry facilitates effective exchange of information about genetic variants by providing globally unique variant identifiers. In addition, the Registry facilitates aggregation of information about variants by enabling instantaneous on-demand registration of new variants and of the links to additional information in external sources about each variant. The Registry is accessible at http://reg.clinicalgenome.org via a web interface and programmatically via well-documented HTTP REST-APIs.
Genetic database software as medical devices
- Pages: 1702-1712
- First Published: 11 October 2018
This article provides a primer on medical device regulation of software that interprets and exchanges genomic data. We compare tests for determining if software qualifies as a medical device across the United States, Europe, and Canada, as well as risk classifications and regulatory controls. Medical device regulation of both software and genetic tools remains uncertain, raising competing concerns: insufficient regulation allows low quality outputs to undermine patient care, while excessive regulation stifles development of innovative tools delivering precision medicine.
The clinical imperative for inclusivity: Race, ethnicity, and ancestry (REA) in genomics
- Pages: 1713-1720
- First Published: 11 October 2018

The Ancestry and Diversity Working Group of the Clinical Genome Resource (ClinGen) presents the results of quantitative and qualitative analyses about race, ethnicity, and ancestry (REA) in clinical genomics. Our findings show great heterogeneity across clinical laboratories in the way race and ethnicity are reported on requisition forms and recommend that standard methods be developed and put into practice through future collaborations. We also demonstrate disparities in the amount of information available for variants at clinically relevant sites across populations.
Adapting crowdsourced clinical cancer curation in CIViC to the ClinGen minimum variant level data community-driven standards
- Pages: 1721-1732
- First Published: 11 October 2018
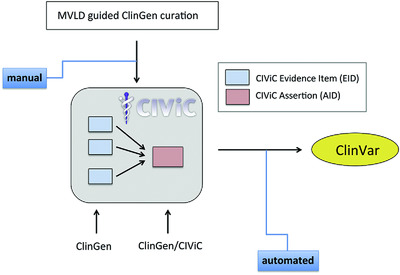
Increasing use of next generation sequencing in cancer generates large volumes of patient variant data for clinical interpretation. Complex intergroup biocuration can help address this problem and includes coordination of group efforts and harmonization of variant formatting. This work describes generation and implementation of such a workflow for collaborative somatic variant curation by ClinGen Somatic Working Groups and the Clinical Interpretation of Variants in Cancer database (CIViC – www.civicdb.org), along with subsequent automated ClinVar submission.