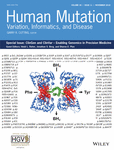
The progression of the ClinGen gene clinical validity classification over time
For the ClinGen/ClinVar Special Issue
Abstract
In order for ClinGen to maintain up-to-date gene-disease clinical validity classifications for use by clinicians and clinical laboratories, an appropriate timeline for reevaluating curated gene-disease associations will need to be determined. To provide guidance on how often a gene-disease association should be recurated, a retrospective analysis of 30 gene curations was performed. Curations were simulated at one-year intervals starting with the year of the first publication to assert disease-causing variants in the gene to observe trends in the classification over time, as well as factors that influenced changes in classification. On average, gene-disease associations spent the least amount of time in the “Moderate” classification before progressing to “Strong” or “Definitive.” In contrast, gene-disease associations that spent five or more years in the “Limited” classification were most likely to remain “Limited” or become “Disputed/Refuted.” Large population datasets contributed to the reclassification of several gene-disease associations from “Limited” to “Disputed/Refuted.” Finally, recent advancements in sequencing technology correlated with an increase in the quantity of case-level evidence that was curated per paper. This study provided a number of key points to consider when determining how often to recurate a gene-disease association.
1 INTRODUCTION
The objective of the NIH-funded Clinical Genome Resource (ClinGen) is to build a central resource that defines the clinical relevance of genes and variants within specific disease entities for use in precision medicine and research. In order to facilitate this effort, the ClinGen Gene Curation Working Group developed a framework for curators to semi-quantitatively define the clinical validity of a gene-disease association (Strande et al., 2017). This process involves the curation and systematic evaluation of peer-reviewed publicly available evidence to assign the strength of a gene-disease association into one of the following clinical validity classifications: Definitive, Strong, Moderate, Limited, No Reported Evidence, or Conflicting Evidence Reported.
As a resource for use by clinicians and clinical laboratories, it is important for the gene curation activities of ClinGen to reflect up-to-date information for gene-disease associations. Thus, the need for reassessment of clinical validity classifications will be crucial, as additional information and evidence is published in the literature. This study examines how the clinical validity classification of a number of genes has changed over time and the factors that influenced those changes. By identifying those variables and trends in the clinical validity classifications over time, we can provide guidance for reevaluating curated gene-disease associations.
2 METHODS
A retrospective analysis of clinical validity classifications was performed on 22 gene-disease associations that were previously assigned a clinical validity classification using the ClinGen framework (Strande et al., 2017) (Table 1). In order to identify information about the gene-disease association as it accumulated, PubMed searches were performed, limited to one-year intervals beginning with the first publication to assert disease-causing variants in a gene. For the 22 gene-disease associations that were chosen, the year of first assertion ranged from 1991 to 2016. The information gathered included genetic (case reports, segregation data, case-control studies), experimental, and contradictory evidence. Using the ClinGen gene curation framework Standard Operating Procedures (SOP), Version 5 (clinicalgenome.org), curators simulated clinical validity assessments at one year-increments. A “Limited” classification scored up to 6 total points, a “Moderate” classification scored 7–11 total points, a “Strong” classification scored 12–18 total points, and a “Definitive” classification scored 12–18 points with replication over time (> 3 years). A “Disputed/Refuted” classification represents those gene-disease associations in which conflicting evidence for the role of the gene in the disease was reported.
| HGNC gene symbol | Disease (inheritance) | Orphanet ID, OMIM phenotype | Disease prevalencea | Year of 1st assertion in the literature (total years curated) | Expert reviewed classification (points) | Notes |
|---|---|---|---|---|---|---|
| KLHL24 | Epidermolysis bullosa simplex (AD) | ORPHA N/A OMIM #617294 | 1:30,000–1:50,000 (Pfender & Bruckner, 1998) | 2016 (1) (Lin et al.) | Strongc (12 GE, 4 EE) | EXPH5, KRT5, KRT14, TGM5 account for ∼81% of cases (Pfender & Bruckner, 1998). |
| SERPINB8 | Exfoliative ichthyosis (AR) | ORPHA 289586 OMIM #617115 | N/A | 2016 (1) (Pigors et al.) | Limitedc (1.75 GE, 1 EE) | |
| ATF6 | Achromatopsia (AR) | ORPHA 49382 OMIM #616517 | < 1:30,000 (Kohl et al., 2004) | 2015 (2) (Kohl et al.) | Strongd (12 GE, 2 EE) | Six genes are associated with the disease. Variants in ATF6 have been found in at least 12 families in the literature (Kohl et al., 2004). |
| XRCC4 | Hereditary colorectal cancer (AD) | ORPHA N/A OMIM N/A | 5–10% of CRC cases are hereditary (Lynch & de la Chapelle, 2003) | 2015 (3) (Esteban-Jurado et al.) | Limitede (1.5 GE, 0 EE) | XRCC4 is also involved in short stature, microcephaly, and endocrine dysfunction (OMIM #616541). |
| LBR | Anadysplasia-like, Spondylometaphyseal Dysplasia (AR) | ORPHA 448267 OMIM N/Ab | < 1:1,000,000 (Orphanet) | 2013 (4) (Borovik et al.) | Moderated (4 GE, 2.5 EE) | |
| ABCC9 | Cantú syndrome (AD) | ORPHA 1517 OMIM #239850 | N/A | 2012 (5) (van Bon et al.) | Definitivec (12 GE, 1 EE, r/t) | Variants in ABCC9 account for ∼28/35 cases in the literature (Grange et al., 2014). |
| WRAP53 | Dyskeratosis congenita (AR) | ORPHA 1775 OMIM #613988 | 1–9:1,000,000 (Orphanet) | 2011 (7) (Zhong et al.) | Moderated (3.5 GE, 6 EE) | Eleven genes are associated with the disease. WRAP53 variants account for < 1% of cases (Savage, 2009). |
| KLF10 | Hypertrophic cardiomyopathy (AD) | ORPHA 217569 OMIM N/A | 1:500 (Cirino & Ho, 2008) | 2012 (7) (Bos et al.) | Limitedf (1.25 GE, 4.5 EE) | >16 genes are associated with HCM. KLF10 variants account for < 1% of cases (Cirino & Ho, 2008). |
| PDZD7 | Sensorinerual hearing loss (AR) | ORPHA 90636 OMIM #618003 | 1:1,000–1:700 (Orphanet) | 2009 (8) (Schneider et al.) | Definitiveg (12 GE, 4 EE, r/t) | 60–80% of cases are of genetic origin. ∼86 causative genes for this disease have been identified (Orphanet). |
| NHP2 | Dyskeratosis congenita (AR) | ORPHA 1775 OMIM #613987 | 1–9:1,000,000 (Orphanet) | 2008 (9) (Vulliamy et al.) | Limitedd (2 GE, 4 EE) | Eleven genes are associated with the disease. NHP2 variants account for < 1% of cases (Savage, 2009). |
| JPH2 | Hypertrophic cardiomyopathy (AD) | ORPHA 217569 OMIM #613873 | 1:500 (Cirino & Ho, 2008) | 2007 (10) (Landstrom et al.) | Moderatef (4.2 GE, 4.5 EE) | >16 genes are associated with HCM. JPH2 variants account for < 1% of cases (Cirino & Ho, 2008). |
| AKAP9 | Long QT syndrome (AD) | ORPHA 101016 OMIM #611820 | 1–5:10,000 (Orphanet) | 2007 (10) (Chen et al.) | Limitedd (0.5 GE, 1.5 EE) | Fifteen genes are associated with LQTS. Variants in AKAP9 account for < 1% of cases (Alders et al., 2003). |
| RPS24 | Diamond-Blackfan anemia (AD) | ORPHA 124 OMIM #610629 | 1:100,000–1:200,000 (Clinton & Gazda, 2009) | 2006 (11) (Gazda et al.) | Definitived (10.5 GE, 3.5 EE, r/t) | ∼2% of cases are attributed to variants in RPS24 (Clinton & Gazda, 2009). |
| TMPO | Dilated cardiomyopathy (AD) | ORPHA 154 OMIM N/A | 1:250 (Hershberger & Morales, 2007) | 2005 (12) (Taylor et al.) | Refutedd | Only 1 proband with a TMPO variant reported in the literature, which was later refuted (Strande et al., 2017). |
| TCAP | Hypertrophic cardiomyopathy (AD) | ORPHA 217569 OMIM #607487 | 1:500 (Cirino & Ho, 2008) | 2004 (13) (Hayashi et al.) | Limitedf (0.2 GE, 1 EE) | >16 genes are associated with HCM. TCAP variants account for < 1% of cases (Cirino & Ho, 2008). |
| MYO1A | Sensorineural hearing loss (AD) | ORPHA 90635 OMIM N/A | 2003 (14) (Donaudy et al.) | Refutedg | ||
| COX15 | Leigh syndrome (AR) | ORPHA 255241/1561 OMIM #256000/615119 | 1:40,000 (Rahman & Thorburn, 2015) | 2003 (15) (Antonicka, et al.) | Strongf,h (8 GE, 5 EE) | <5% of complex IV-deficient Leigh syndrome is caused by variants in COX15 (Rahman & Thorburn, 2015). |
| USH1C | Usher syndrome type 1C (AR) | ORPHA 231169 OMIM #276904 | 1–9:100,000 (Orphanet) | 2000 (17) (Verpy et al.) | Definitiveg (12 GE, 6 EE, r/t) | Six genes are associated with the disease. 1–15% of cases are attributed to variants in USH1C (Lentz & Keats, 1999). |
| SMAD4 | Juvenile polyposis syndrome (AD) | ORPHA 329971 OMIM #174900 | 1:16,000–1:100,000 (Haidle & Howe, 2003) | 1998 (19) (Howe et al.) | Definitivee (12 GE, 6 EE, r/t) | ∼27% of cases are attributed to variants in SMAD4 (Haidle & Howe, 2003. |
| PMS1 | Lynch syndrome (AD) | ORPHA 144 OMIM N/A | 1:440 (Kohlmann & Gruber, 2004) | 1994 (23) (Nicolaides et al.) | Refutede | Variants in MLH1, MSH2, MSH6, PMS2 and EPCAM account for most cases (Kohlmann & Gruber, 2004). |
| TPM1 | Hypertrophic cardiomyopathy (AD) | ORPHA 217569OMIM #115196 | 1:500 (Cirino & Ho, 2008) | 1994 (23) (Thierfelder et al.) | Definitivef (12 GE, 6 EE, r/t) | ∼2% of HCM cases are caused by variants in TPM1 (Cirino & Ho, 2008). |
| GAA | Pompe disease (AR) | ORPHA 365 OMIM #232300 | 1:14,000–1:100,000 (Leslie & Bailey, 2007) | 1991 (26) (Zhong et al. 2011) | Definitivec (12 GE, 6 EE, r/t) | GAA is the only gene associated with Pompe disease (Leslie & Bailey, 2007). |
- AD, autosomal dominant; AR, autosomal recessive; N/A, not applicable, r/t, replication over time; GE, genetic evidence; EE, experimental evidence.
- a Overall disease prevalence, not specific to gene.
- b An OMIM entry for Pelger–Huet anomaly with mild skeletal anomalies (OMIM #618019) has been created since the original curation was performed (Strande et al., 2017) and may represent the most appropriate disease entity. This should be considered and updated upon reanalysis of the gene-disease association.
- Classifications approved by:
- c Jonathan Berg, MD, PhD, ClinGen.
- d Reported in Strande et al., 2017.
- e ClinGen Colon Cancer and Polypsosis Gene Curation Expert Panel.
- f ClinGen Hypertrophic Cardiomyopathy Gene Curation Expert Panel.
- g ClinGen Hereditary Hearing Gene Curation Expert Panel.
- h Heather Baudet, MD, PhD, ClinGen.
Population databases—the Exome Aggregation Consortium (ExAC; exac.broadinstitute.org) and the Genome Aggregation Consortium (gnomAD; gnomad.broadinstitute.org) (Lek et al., 2016)—were utilized to assess variant pathogenicity, but were only taken into account for scoring metrics based on the point in time of their public release (October, 2014 for ExAC and October, 2016 for gnomAD) in order to observe how these databases influence the interpretation of a variant's pathogenicity. The allele frequency at which a variant was considered benign was determined by the disease domain experts for each gene-disease curation.
To assess how the clinical validity classifications would evolve in more recently asserted gene-disease associations, eight novel gene-disease associations reported in 2013 in the American Journal of Human Genetics, volume 93 (issues 1 and 2) were selected for analysis (Table 2). Curations were simulated at one-year increments from 2013 to 2016.
| HGNC gene symbol | Disease (inheritance) | Orphanet ID, OMIM phenotype | Disease prevalencea | Expert reviewed classification (points) | Notes | Reference |
|---|---|---|---|---|---|---|
| AQP5 | Palmoplantar keratoderma, Bothnian type (AD) | ORPHA 2337 OMIM #600231 | 1:40,000, 0.3–0.55% in northern Sweden (Orphanet) | Moderateb (7.25 GE, 3.5 EE) | Blaydon et al., 2013 | |
| ELAC2 | Combined oxidative phosphorylation deficiency (AR) | ORPHA 369913 OMIM #615440 | N/A | Moderateb (6 GE, 4 EE) | >20 genes are involved in complex I deficiency (OMIM #252010). | Haack et al., 2013 |
| KLHL40 | Nemaline myopathy, severe congenital (AR) | ORPHA 171430 OMIM #615348 | 1:50,000 (North & Ryan, 2002) | Definitiveb (12 GE, 6 EE, r/t) | Classified as moderate in 2013, strong in 2014, definitive in 2016 due to r/t. There are ∼10 genes involved in the disease (North & Ryan, 2002). | Ravenscroft et al., 2013 |
| LRPAP1 | Myopia (AR) | ORPHA 98619 OMIM #615431 | ∼25% (Aldamesh, 2013) | Moderateb (9.5 GE, 1.5 EE) | Linkage analysis has identified at least 13 loci for high myopia (Jiang et al., 2014). | Aldahmesh et al., 2013 |
| PIK3R1 | SHORT syndrome (AD) | ORPHA 3163 OMIM #269880 | <50 cases in the literature (Innes & Dyment, 2014) | Definitiveb (12 GE, 3 EE, r/t) | Upgraded to Definitive in 2016 due to replication over time. PIK3R1 is the only gene currently associated with the disease (Innes & Dyment, 2014). | Chudasama et al., 2013; Dyment et al., 2013; Thauvin-Robinet et al., 2013 |
| RAB28 | Cone-rod dystrophy (AR) | ORPHA 1872 OMIM #615374 | 1:30,000–1:40,000 (Roosing et al., 2013) | Limitedb (3.75 GE, 1.5 EE) | Seven genes have been previously implicated in the AR form of the disease (Roosing et al., 2013). | Roosing et al., 2013 |
| ARL2BP | Retinitis pigmentosa (AR) | ORPHA 791 OMIM #268000 | 1–5:10,000 (Orphanet) | Limitedb (4.5 GE, 1.5 EE) | AR cases make up 5–20% of cases. There are more than 50 genes associated with the AR condition (Fahim et al., 2000). | Davidson et al., 2013 |
| CTCF | Intellectual disability (AD) | ORPHA 363611 OMIM #615502 | 1:1,000,000 (Orphanet) | Moderatec (4.5 GE, 6 EE) | Gregor et al., 2013 |
- AD, autosomal dominant; AR, autosomal recessive; N/A, not applicable; r/t, replication over time; GE, genetic evidence; EE, experimental evidence.
- a Overall disease prevalence, not specific to gene.
- Classifications approved by:
- b Jonathan Berg, MD, PhD, ClinGen.
- c Christian Schaaf, MD, PhD, FACMG, ClinGen.
Statistical analysis was performed using an unpaired t-test (GraphPad). All current gene-disease validity summaries can be found on the ClinGen website (www.clinicalgenome.org) by searching the HGNC gene symbol provided in Tables 1 and 2.
3 RESULTS
In order to observe any trends in clinical validity classifications over time, a diverse selection of 22 curated gene-disease associations (Table 1) were evaluated at one-year intervals using the ClinGen gene curation framework, starting with the year of the first publication to assert disease-causing variants in the gene of interest (Figure 1). Of the 22 gene-disease associations, six are currently classified as “Limited,” three are “Moderate,” three are “Strong,” seven are “Definitive” and three are “Refuted.” The gene-disease associations were chosen to encompass a range of clinical domains, inheritance patterns, pathologies, molecular mechanisms, and time at first assertion (1991–2016) and were selected from available curations that had been performed by ClinGen Gene Curation Expert Panels.
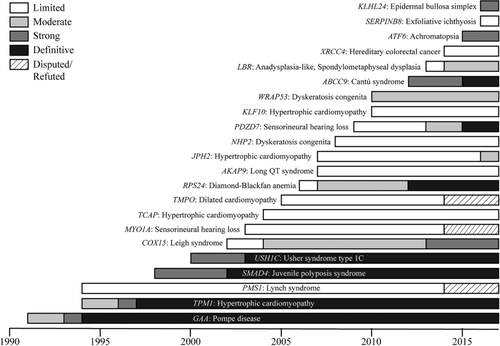
On average, over the entire lifetime of a gene-disease association (from first assertion to the latest assessment for this study, 2017), gene-disease pairs tended to spend a higher percentage of time at the “Limited” classification (71.4 ± 34.9%) compared to “Moderate” (41.5 ± 34.6%) (Figure 2A). Given that the point range for the “Moderate” classification is four points (7–11), compared to “Limited” and “Strong/Definitive” that are 5.9 (0.1–6) and six points (12–18), respectively, it is logical that gene-disease associations would spend the least amount of time in the “Moderate” classification. Consistent with this observation, we found that 71% of gene-disease associations that spent more than a year at the “Moderate” classification progressed to “Strong” (1/7) or “Definitive” (4/7) by 2017. Of note, JPH2: hypertrophic cardiomyopathy was excluded from this analysis as it had only reached the “Moderate” classification within the last year of the curation time frame, and thus was not subject to the analysis. Only 2/7 gene-disease associations remain at “Moderate” (Figure 2B), suggesting that these associations may spend a fractional amount of time at “Moderate” before advancing to a new classification. Furthermore, it should be noted that we did not encounter a gene-disease association that achieved a “Moderate” classification that then moved to a “Disputed” or “Refuted” classification, further supporting the suggestion that once a gene-disease association meets a classification of “Moderate” it will most likely advance to “Strong” or “Definitive.”
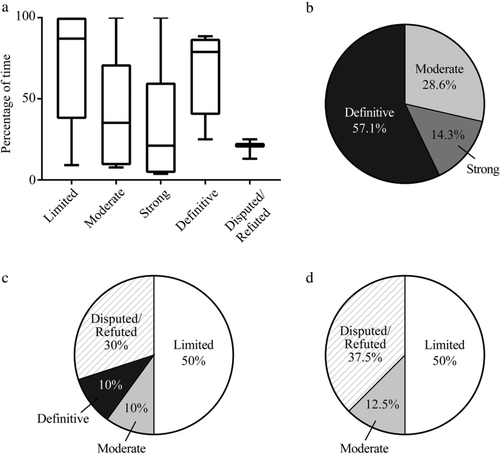
In assessing gene-disease associations with a “Limited” classification, we found that out of the ten genes that spent at least three or more years at “Limited,” 50% of them currently remain at “Limited.” Only 2/10 progressed to a higher classification: one each to “Moderate” and “Definitive,” while 3/10 became “Disputed/Refuted” (Figure 2C). This observation is consistent with the finding that gene-disease associations spend more time at the “Limited” classification, compared to “Moderate” and “Strong” (Figure 2A). This trend was even more significant when we looked at gene-disease associations that spent at least five or more years at “Limited”: only 1/8 progressed to a higher classification, while 4/8 stayed at “Limited” and 3/8 became “Disputed/Refuted” (Figure 2D). It is interesting to note that gene-disease associations that began as “Limited” and were reclassified as “Disputed/Refuted” only reached a maximum of two points in genetic and experimental evidence before conflicting evidence was reported (TMPO: dilated cardiomyopathy, MYO1A: sensorineural hearing loss, and PMS1: Lynch syndrome). In contrast, “Limited” gene-disease associations that progressed to a higher classification (LBR: anadysplasia-like, spondylometaphyseal dysplasia, PDZD7: sensorineural hearing loss, JPH2: hypertrophic cardiomyopathy, RPS24: Diamond-Blackfan anemia, and COX15: Leigh syndrome) initially scored more than two points at the time of the first assertion. These findings suggest that gene-disease associations at the low end of the “Limited” point range (< 2 points) are more likely to remain “Limited” or become “Disputed/Refuted” than gene-disease associations that initially scored in mid to upper “Limited” point range (2–6 points).
3.1 Disputed/refuted gene-disease associations
Three out of 22 genes analyzed spent between 9 and 20 years at “Limited” before being reclassified as “Disputed” or “Refuted” between 2013 and 2014 (Figure 1). For each of these gene-disease associations, the disputing or refuting factor that contributed most to the reclassification was the high maximum allele frequencies recorded for the asserted disease-causing variant(s) in the healthy control populations, as assessed by the use of ExAC and gnomAD. For the “Disputed” or “Refuted” categories, it is worth noting that the classifications are based on a specific claim of a monogenic gene-disease association. Thus, any dispute/refute is specific to that association and does not exclude other claims as being valid, nor does it suggest that the gene is not involved in any other disease by means of a different mechanism and/or mode of inheritance.
In 2005, TMPO: dilated cardiomyopathy was classified as “Limited” based on one case report of a proband with a missense variant (Taylor et al., 2005). However, with the public release of the ExAC database in 2014, this variant was reported at an overall frequency of 0.01508 in the population and was found in the homozygous state in 141 individuals, suggesting the variant is benign. Therefore, TMPO: dilated cardiomyopathy was classified as “Refuted” (Strande et al., 2017).
PMS1: Lynch syndrome was originally given a classification of “Limited” in 1994 based on the report of a proband harboring a variant that resulted in exon skipping (Nicolaides et al., 1994). In 2001, when the patient's genome was further analyzed, it was demonstrated that she also carried a pathogenic mutation in MSH2 (which is also associated with Lynch syndrome) that segregated with disease in the family, indicating that the MSH2 variant was the probable causative variant (Liu et al., 2001). There was only one additional report in 1999 of a proband carrying two missense variants in PMS1 (Wang et al., 1999), which were later found at an overall frequency of 0.04513 and 0.04594 in ExAC, each with 11 homozygotes, suggesting that they are both benign. This led to a “Refuted” classification for PMS1: Lynch syndrome (ClinGen Colon Cancer and Polyposis Gene Curation Expert Panel, clinicalgenome.org).
Finally, MYO1A: Sensorineural hearing loss was classified as “Limited” in 2003 (Donaudy et al., 2003) based on the identification of six missense variants, one in-frame deletion, and one nonsense variant in MYO1A in probands with hearing loss. However, four of the missense variants were later found at an allele frequency greater than 0.5% in gnomAD, and the nonsense variant was found to be inherited from an unaffected mother. A 2014 report identified alternate genetic causes of hearing loss in two probands with novel variants in MYO1A and one proband that was originally reported by Donaudy et al. (2003) (Eisenberger et al., 2014). Finally, a 2017 report identified 12 individuals without hearing loss that harbored either a loss-of-function variant in MYO1A or one of the variants that had been reported by Donaudy et al. (2003) (Patton et al., 2016). Each of these factors led to a final classification of “Refuted” (ClinGen Hereditary Hearing Loss Gene Curation Expert Panel, clinicalgenome.org).
3.2 Impact of new sequencing technologies on clinical validity classifications
In recent years, advancements in sequencing technologies, such as next-generation sequencing (NGS) and whole exome sequencing (WES), have allowed for rapid sequencing of DNA from large numbers of individuals at a fraction of the cost, which has led to increased use of genetic testing in healthcare and research to facilitate genetic variant identification. Additionally, these technologies have led to the development of large genetic population datasets, such as ExAC, which was publicly released in October, 2014 (Lek et al., 2016). In order to determine how these recent advancements in genomics affect the discovery and clinical validity of novel gene-disease associations, we simulated clinical validity assessments of eight novel gene-disease associations reported in 2013 in the American Journal of Human Genetics at one-year intervals from 2013 through 2016 (Table 2).
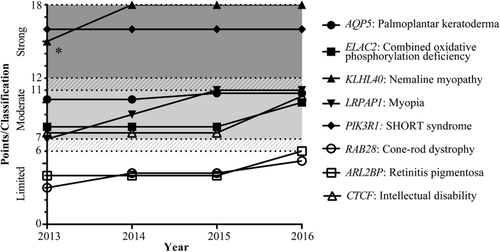
Interestingly, besides advancing from “Strong” to “Definitive,” only one gene-disease association changed classification during the 2013–2016 time period-KLHL40: nemaline myopathy (Figure 3). Although the total points for the initial gene-disease association in 2013 were within the range for a “Strong” classification, it was constrained to a “Moderate” level because there was only one study with evidence to support the role of KLHL40 in nemaline myopathy (Ravenscroft et al., 2013). According to the ClinGen Clinical Validity Classification system, a gene-disease association is classified as “Strong” if the role of the gene in the disease has been demonstrated independently in at least two separate studies. As additional studies were reported in 2014, it was upgraded to “Strong.” Conversely, PIK3R1: SHORT syndrome reached the “Strong” classification in the first year (2013) because three independent studies implicating the gene's role in disease were published in the same issue (Chudasama et al., 2013; Dyment et al., 2013; Thauvin-Robinet et al., 2013). Both KLHL40: nemaline myopathy and PIK3R1: SHORT syndrome reached a final classification of “Definitive” in 2016 due to replication over time.
In order to further investigate how the next generation of sequencing technologies may have impacted the clinical validity classification over time, we focused our analysis to the ten “Definitive” gene-disease associations (Tables 1 and 2, Figures 1 and 3). The ten gene-disease pairs were divided into two groups: those that were discovered using traditional methods (linkage analysis, direct sequencing) prior to 2010 (GAA: Pompe disease, TPM1: hypertrophic cardiomyopathy, SMAD4: juvenile polyposis syndrome, USH1C: Usher syndrome type 1C, RPS24: Diamond-Blackfan anemia, and PDZD7: sensorineural hearing loss) and those that were discovered using modern genomic technology (WES) after 2010 (ABCC9: Cantu syndrome, KLHL40: nemaline myopathy, and PIK3R1: SHORT syndrome).
There was no significant difference in the average amount of time it took for the gene-disease associations to reach “Definitive” between the pre-2010, traditional methods group (4.33 ± 1.37 years) versus the post-2010, WES group (3.0 ± 0 years, P = 0.1465) (Figure 4A). However, there was a significant increase in the average number of probands curated per paper for the post-2010, WES group (1.98 ± 0.55 vs. 8.5 ± 3.29, P = 0.0015) (Figure 4B), suggesting that next-generation sequencing technologies may allow researchers to analyze a greater number of cases, while those gene-disease associations that were discovered using traditional methods relied on more papers to reach “Definitive” status. In fact, this trend was observed when we looked at the number of papers curated for both groups (Figure 4C), though it did not reach significance, which is likely due to a small sample size. For the pre-2010 group, an average of 9.7 ± 3.7 papers were curated to reach “Definitive” status versus 5.0 ± 1.0 papers curated for the post-2010 group (P = 0.0742).

4 DISCUSSION
As the number of completed gene curations continues to grow, ClinGen gene curation groups will need to determine how often gene-disease associations will be reevaluated in order to stay up-to-date with new information as it becomes available in the literature. Analysis of the 30 gene-disease associations discussed here can begin to provide insight into how the clinical validity classification changes over time. This analysis is not only relevant to ClinGen curation activities, but can also be applied by any groups offering diagnostic testing to determine how often genes on panels are assessed. A systematic approach for reanalysis of genes on diagnostic testing panels will help to ensure the most accurate results are reported.
The first thing to consider when determining how often to reevaluate a gene-disease association is the initial clinical validity classification. In effect, the “Limited” classification was intended to represent a relatively weak single publication with few cases and little supporting functional evidence, while the “Moderate” classification was intended to represent a stronger initial publication providing more cases and ample functional evidence supporting the gene's role in the disease. A “Strong” classification implies additional independent corroboration of an initial gene-disease association, increasing both the case-level and gene-level evidence.
There are several reasons why gene-disease associations may stay at “Limited” for a longer period of time. First, the disease may be rare, in which case there are very few probands reported in the literature. It is important to note, however, that with the ClinGen framework, it is possible for even rare gene-disease associations to reach a “Strong” or “Definitive” classification based on the strength of available variant evidence and supporting experimental evidence. Secondly, the gene of interest may be only one of many genes known to cause the same disease and therefore variants in that gene only account for a small number of cases. For example, TCAP, AKAP9, NHP2, and KLF10 account for fewer than 1% of cases of their respective diseases, which are caused by variants in more than ten genes each (see notes on Table 1). Finally, a gene-disease association may spend a greater time at “Limited” due to disparities in research and funding for different clinical domains. For these reasons, a gene-disease association may remain at “Limited” for several years, but with the availability of technologies such as WES, and resources such as GeneMatcher (www.genematcher.org) and GenomeConnect (www.genomeconnect.org) that allow clinicians and researchers who are interested in the same gene to connect, it may eventually progress to a new classification with time.
When determining how often to reevaluate a “Limited” gene-disease association, it is appropriate to consider the length of time spent at “Limited” and the total number of points. Gene-disease associations with greater than five years at “Limited” and points within the lower range of the “Limited” classification (e.g. < 2 points) should be reevaluated less frequently than newly discovered “Limited” gene-disease associations that have points in the upper range of the “Limited” classification. For example, out of the 30 gene-disease associations examined here, there are several “Limited” gene-disease associations at the higher end of the classification point range, and thus are likely to change classification with one or two additional studies, including: KLF10: hypertrophic cardiomyopathy, NHP2: dyskeratosis congenita, RAB28: cone-rod dystrophy, and ARL2BP: retinitis pigmentosa (Tables 1 and 2). Conversely, “Limited” gene-disease associations, such as AKAP9: long QT syndrome and TCAP: hypertrophic cardiomyopathy, that have fewer than two points and have remained “Limited” for more than 10 years (Table 1, Figure 1) should be reevaluated less frequently, and importantly they should be monitored for newly published data that could dispute or refute the gene-disease association.
Not surprisingly, gene-disease associations tend to spend less time in the “Moderate” category compared to “Limited.” This is likely due to the small point range for this category compared to “Limited” and “Strong/Definitive” and the fact that any corroborating publications would be likely to push the supporting evidence into the “Strong” category. Similar to gene-disease associations at the high end of the point range for “Limited,” curation groups may want to reevaluate “Moderate” associations more frequently, and automated systems for alerting curation groups to new publications about the gene or disease would help to keep categorizations up to date. Gene-disease associations also spend very little time at “Strong” due to the fact that they can reach “Definitive” once the association has been upheld for at least three years (Strande et al., 2017). “Strong” gene-disease associations would therefore only need to be reevaluated once they reach the 3-year mark for replication over time.
Given the availability of more advanced genomic sequencing technologies, large population databases, and resources like GeneMatcher and GenomeConnect, it was surprising that most of the novel gene-disease associations from 2013 did not change classification between 2013 and 2016 (Figure 3). It was observed that newer gene-disease associations tend to have more probands per paper (Figure 4B), perhaps due to the use of relatively inexpensive genomic sequencing technologies, match-making and global communication that enables groups to gather cohorts of patients more easily, or the journal's selectivity for the number of probands in a publication, Therefore, the absence of classification changes between 2013 and 2016 could be due to the rarity of the disease, or that variants in the gene only account for a small fraction of cases. However, it is important to note that all four gene-disease associations that fell within the “Moderate” point range in 2013 (LRPAP1: myopaia, AQP5: palmoplantar keratoderma, ELAC2: combined oxidative phosphorylation deficiency, and CTCF: intellectual disability) were within two points of the “Strong/Definitive” classification by 2016. Similarly, both gene-disease associations that fell within the “Limited” point range in 2013 (RAB28: cone-rod dystrophy and ARL2BP: retinitis pigmentosa) were within one point of the “Moderate” classification by 2016, suggesting that they might change classification with an additional publication or two.
In summary, the results from this retrospective study provide several key points to consider when determining how often to reevaluate a gene-disease association. First, “Limited” classifications should be considered carefully. Gene-disease associations that have been at the low end of the “Limited” point range (e.g. < 2 points) for an extended period of time (e.g. > 5 years) and have not been disputed/refuted with the release of ExAC or gnomAD, may not need to be evaluated as frequently as those at the higher end of the “Limited” point range and those that have been discovered more recently. Low “Limited” gene-disease associations may only need to be evaluated every three to five years, while high “Limited” gene-disease associations every one to two years. Eventually, consensus groups may need to reassess the original evidence and determine whether failure to replicate the initial finding constitutes a justification for disputing the gene-disease association. “Moderate” gene-disease associations should be reevaluated more frequently, possibly every one to two years, given the small point range for the category. Finally, “Strong” gene-disease associations should be re-evaluated at the three-year mark to reflect replication over time, allowing the association to reach the “Definitive” classification. These recommendations are based on our initial review. However, reanalysis and adjustment of these time points may be required in the future once more gene-disease associations have been analyzed.
Overall, this study has provided insight into the process of biocuration and has given key consideration points in determining the most effective reevaluation process for gene-disease association classifications.
ACKNOWLEDGMENTS
We wish to thank NHGRI for financial support (U41HG009650, U41HG006834 and U01HG007437) and Erin Riggs, Ahmad Abou Tayoun, Heather Baudet, Christian Schaff, and the following ClinGen groups for their contributions: Gene Curation Working Group, Hypertrophic Cardiomyopathy Gene Curation Expert Panel, Colon Cancer and Polyposis Gene Curation Expert Panel, and Hereditary Hearing Loss Gene Curation Expert Panel.
ClinGen Gene Curation SOP, https://www.clinicalgenome.org/curation-activities/gene-disease-validity/educational-and-training-materials/standard-operating-procedures/
OMIM, https://omim.org/