Human Mutation provides a unique forum for the exchange of ideas, methods, and applications of interest to molecular, human, and medical geneticists in academic, industrial, and clinical research settings worldwide.
Journal Metrics
- 8.6CiteScore
- 3.7Journal Impact Factor
- 17%Acceptance rate
- 84 days Submission to first decision
Articles
Prognostic Value of Ubiquitination‐Related Genes in Ovarian Cancer and Their Correlation With Tumor Immunity
- 15 July 2025
Bayesian Optimization–Enhanced Machine Learning for Osteosarcoma Risk Stratification Based on Sphingolipid Metabolism
- 11 July 2025
Immune Dysregulation and Prognosis in Sepsis: Insights From a Posttranslational Perspective
- 8 July 2025
Adaptation of ACMG/AMP Guidelines for Clinical Classification of BMPR2 Variants in Pulmonary Arterial Hypertension Resolves Variants of Unclear Pathogenicity in ClinVar
- 7 July 2025
The following is a list of the most cited articles based on citations published in the last three years, according to CrossRef.
GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene
- 928-930
- 29 July 2015
HGVS Recommendations for the Description of Sequence Variants: 2016 Update
- 564-569
- 2 March 2016
Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion
- 1517-1524
- 7 September 2018
Graphical Abstract
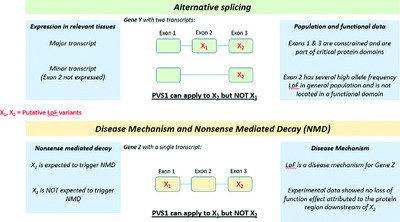
We provide guidance for PVS1 usage that takes into consideration all aspects of putative loss of function (LoF) variants, including type, location, and annotation, and the disease mechanism of the genes they affect. We demonstrate how the combination of these variant and gene attributes can lead to varied PVS1 strength levels. Finally, we evaluate the refined criterion using > 50 LoF variants in several genes and diseases.
Variant interpretation using population databases: Lessons from gnomAD
- 1012-1030
- 2 December 2021
Graphical Abstract
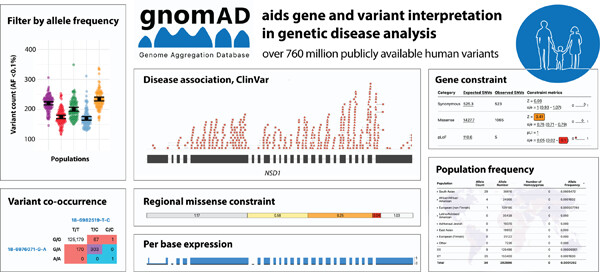
Reference population databases are critical in the interpretation of genomic variation for diagnosing rare disease, and supports the discovery of new disease–gene relationships. This review provides guidance for using the Genome Aggregation Database (gnomAD) browser and key features like allele frequency, per-base expression levels, constraint scores, and variant co-occurrence, for variant and gene interpretation in clinical and research analysis.