Adaptation of ACMG/AMP Guidelines for Clinical Classification of BMPR2 Variants in Pulmonary Arterial Hypertension Resolves Variants of Unclear Pathogenicity in ClinVar
Abstract
Pulmonary arterial hypertension (PAH) is a rare disease that can be caused by pathogenic variants, most frequently in the bone Morphogenetic Protein Receptor Type 2 (BMPR2) gene. We formed a ClinGen variant curation expert panel to devise guidelines for the clinical interpretation of BMPR2 variants identified in PAH patients. The general ACMG/AMP variant classification criteria were refined for PAH and adapted to BMPR2 following ClinGen procedures. Subsequently, these specifications were tested independently by three members of the curation expert panel on 28 representative BMPR2 variants selected from ClinVar and then presented and discussed in the plenum. Application of the final BMPR2 variant specifications resolved six of nine variants (66%) where multiple ClinVar classifications included a variant of uncertain significance, with all six being reclassified as Benign or Likely Benign. Four splice site variants underwent clinically consequential reclassification based on the presence or absence of supporting mRNA splicing data. These variant specifications provide an international framework and a valuable tool for BMPR2 variant classification that can be applied to increase confidence and consistency in BMPR2 interpretation for diagnostic laboratories, clinical providers, and patients.
1. Introduction
Pulmonary arterial hypertension (PAH; OMIM #178600) is a rare disease with a prevalence of 15–50 cases per one million individuals [1, 2]. Symptoms of PAH are nonspecific including shortness of breath particularly during exertion, palpitations, and, in advanced cases, edema, chest pain, and dyspnea at rest. While noninvasive assessments such as echocardiography, electrocardiography, laboratory tests, pulmonary function tests, and cardiopulmonary exercise testing can support a diagnosis of PAH, the definitive diagnosis is made by right heart catheterization. PAH is hemodynamically defined by an elevated mean pulmonary artery pressure > 20 mmHg, pulmonary vascular resistance > 2 Wood units, and a pulmonary artery wedge pressure ≤ 15 mmHg [2]. PAH is currently classified as idiopathic (IPAH), heritable (HPAH), or associated with other diseases or exposures, including connective tissue disease, congenital heart disease, HIV infection, or drugs and toxins. By definition, patients with HPAH have a positive family history and/or carry a pathogenic variant in a definitive PAH risk gene. Thus, apparently idiopathic patients may be reclassified as HPAH following genetic analysis.
The first gene identified as causative for PAH encodes the Bone Morphogenetic Protein Receptor Type 2 (BMPR2; HGNC: 1078) [3, 4], and it remains the gene with the highest frequency of Pathogenic variants in HPAH patients [5–7]. BMPR2 is a cell surface receptor and part of the Transforming Growth Factor β superfamily responsible for vascular homeostasis of cell proliferation and apoptosis. Heterozygous Pathogenic or Likely Pathogenic (LP) BMPR2 variants lead to PAH in an autosomal dominant manner with reduced penetrance of approximately 15%–40% and show sexual dimorphism, with higher penetrance in females [8].
Subsequently, several additional genes have been implicated in the etiology of IPAH and HPAH, especially with the advent of next-generation sequencing of large cohorts [9–11]. A recent study carried out by our Clinical Genome Resource (ClinGen) pulmonary hypertension gene curation expert panel (PH GCEP) thoroughly characterized 27 genes and classified 12 of them to have a definitive gene–disease relationship with IPAH/HPAH (ACVRL1, ATP13A3, BMPR2, CAV1, EIF2AK4, ENG, GDF2, KCNK3, KDR, SMAD9, SOX17, and TBX4), three to have moderate evidence (ABCC8, GGCX, and TET2), six with limited evidence (AQP1, BMP10, FBLN2, KLF2, KLK1, and PDGFD), and one to have no evidence for the relationship with PAH (TOPBP1) [12]. Notably, four genes in the BMPR2 pathway (BMPR1A, BMPR1B, SMAD1, and SMAD4) were disputed as causative PAH genes [12].
ClinGen, funded by the National Institutes of Health (NIH), serves as a central resource with information on the clinical significance of genes and variants [13]. Curation of genes and variants focuses on gene disease validity, variant pathogenicity, dosage sensitivity, and clinical actionability. ClinGen also offers the opportunity to form expert panels to curate not only genes but also variants [14]. To this end, following our gene curation efforts, we founded a PH variant curation expert panel (VCEP) to adapt the American College of Medical Genetics and Genomics (ACMG)/Association of Molecular Pathology (AMP) guidelines with a focus on BMPR2. We modified the general variant curation guidelines by the ACMG/AMP framework [15] for BMPR2 variants specific to HPAH and/or IPAH. Since a significant number of BMPR2 variants have an unclear impact on gene expression or function we aimed to resolve conflicting classifications to improve the interpretation of genetic test results. We tested our refined specification criteria on a pilot subset of 28 representative BMPR2 variants, the results of which are described herein.
2. Methods
Within the framework of our Genetics Task Force of the International Consortium for Genetic Studies in PAH (PAH-ICON, https://pahicon.com/), we founded a ClinGen Pulmonary Hypertension VCEP with 16 members from nine institutions across the United States and Europe, with representation from six countries (https://www.clinicalgenome.org/affiliation/50071/). Members included a chairperson (WKC), scientific lead/coordinator (CLW), expert reviewers, and biocurators. Expert reviewer and biocurator roles were assigned based on members’ experience, but most members had dual roles. Seven members hold professional certifications in clinical genetics and/or regularly use ACMG/AMP guidelines to classify variants for clinical laboratory case sign-out. Most members had a history of ClinGen activities including curation of PAH gene relationships [12]. All members received additional training in the ClinGen variant curation standard operating procedure V3, the 2015 ACMG/AMP guidelines, the ClinGen sequence variant interpretation working group recommendations for applying ACMG/AMP criteria, sequence variant literature searches, and use of ClinGen’s variant curation interface (VCI) [16]. We followed the ClinGen four-step process (protocol Version 9) to obtain VCEP approval (February 2024) with resulting variant classifications meeting US Food and Drug Administration (FDA) recognition. A flow chart for the VCEP formation and activities is shown in Figure 1.
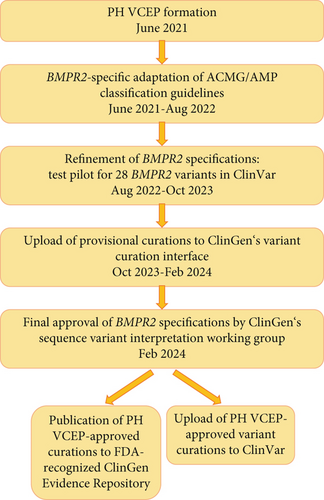
We limited the scope of this work to BMPR2 variants because of the large number of loss-of-function variants, recurrent variants, and the relatively large body of experimental evidence defining critical domains/residues and functional effects of individual variants compared to other PAH risk genes [12]. Monthly virtual meetings were held over a 14-month period (June 2021 to August 2022) to develop variant classification rules for BMPR2 based on the ACMG/AMP guidelines (Figure 1). We then tested our adapted specifications on a pilot group of 28 variants, representing all ACMG classes, including those with conflicting interpretations of pathogenicity. The selected variants included insertions, deletions, 5 ′-untranslated region, missense, splice, nonsense, and synonymous variants. Only BMPR2 variants identified in nonsyndromic HPAH and/or IPAH cases were considered for pilot curation. Thus, one additional variant (c.319T>C p.Ser107Pro) was removed from the pilot curation list as it had been described in a proband with congenital heart disease–associated PAH and was therefore outside the scope of this work. The BMPR2 MANE Select transcript with accession number NM_001204.7 was used as the reference for all variant curations.
We assigned six variants to each curator and teams of three curators per variant. Provisional classifications were presented and discussed at monthly meetings over another 14-month period (August 2022 to October 2023). Modifications of our BMPR2 criteria specifications were made over this period and following guidance of the ClinGen sequence variant interpretation working group. The specifications are available in the ClinGen Criteria Specification Registry (https://cspec.genome.network/cspec/ui/svi/doc/GN125). Final curations were uploaded to the VCI. Following the final PH VCEP approval, variant curations were published to the ClinGen Evidence Repository and submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/?term=bmpr2%255Bgene%255D%26redir=gene). All members disclosed conflicts of interest, and the curation assignments were mindful of conflicts. Our ACMG/AMP specifications are updated periodically. To find the most current information, please visit https://cspec.genome.network.
3. Results
3.1. Development of BMPR2-Specific Variant Curation Criteria
3.1.1. Population Frequency-Based Criteria: PM2, BA1, BS1, and BS2
The population frequency criteria evaluation includes four criteria (Table 1). Based on our BMPR2 variant specifications, we applied PM2 as supporting when the variant was absent from the gnomAD V2.1.1 (controls) and V3.1.2 (controls/biobanks) populations or had an allele frequency below 0.01%. For this, we considered the ancestral subgroup with the highest frequency and at least 1000 allele counts. The 0.01% threshold for allele frequency was defined based on the current prevalence of PAH of 15–50 cases/million individuals and accounting for reduced penetrance [1, 2].
| Pathogenic criteria | ||
|---|---|---|
| Criteria | Criteria description | Specification |
| Very strong criteria | ||
| PVS1 | Null variant in a gene where loss of function is a known mechanism of disease | Use BMPR2-specific PVS1 decision tree (Supporting Information 1: Figure S1) |
| Strong criteria | ||
| PS1 | Same amino acid change as a previously established Pathogenic variant regardless of nucleotide change | Downgrade to PS1_mod if variant is likely pathogenic |
| PS2 | De novo (paternity confirmed) in a patient with the disease and no family history | — |
| PS3 | Well-established in vitro or in vivo functional studies supportive of a damaging effect |
|
| PS4 | The prevalence of the variant in affected individuals is significantly increased compared with the prevalence in controls |
|
| Moderate criteria | ||
| PM1 | Located in a mutational hot spot and/or critical and well-established functional domain | Extracellular domain: p.Cys34, Cys60, Cys66, Cys84, Cys94, Cys99, Cys116, Cys117, Cys118, and Cys123. Kinase domain: p.Gly210, Gly212, Lys230, Glu/Asn245, Asp333, Asn338, Asp351, Gly353, Glu386, Asp405, Gly410, and Arg491. Heterodimerization: p.Asp485, Gln486, Asp487, Ala488, Arg489, Ala490, Arg491, and Leu492 |
| PM2 |
|
|
| PM3 | For recessive disorders, detected in trans with a pathogenic variant | N/A |
| PM4 | Protein length changes due to in-frame deletions/insertions in a nonrepeat region or stop-loss variants | — |
| PM5 | Missense change at an amino acid residue where a different missense change determined to be pathogenic has been seen before | If the second variant is pathogenic, then apply PM5_mod. If the second variant is likely pathogenic, then apply PM5_supp |
| PM6 | Confirmed de novo without confirmation of paternity and maternity | — |
| Supporting criteria | ||
| PP1 |
|
|
| PP2 | Missense variant in a gene that has a low rate of benign missense variation and where missense variants are a common mechanism of disease | N/A |
| PP3 | Multiple lines of computational evidence support a deleterious effect on the gene or gene product | REVEL ≥ 0.75 for missense, no up/downgrading; SpliceAI ≥ 0.2 for noncanonical splice variants. If no REVEL score or SpliceAI prediction available, then CADD ≥ 20 can be used. PP3 can be applied additively with PS3 |
| PP4 | Phenotype specific for disease with single genetic etiology | N/A |
| PP5 | Reputable source recently reports variant as pathogenic, but the evidence is not available to the laboratory to perform an independent evaluation | N/A not to be used |
| Benign criteria | ||
| Criteria | Criteria description | Specification |
| Stand-alone criteria | ||
| BA1 | Allele frequency above X% | 1% |
| Strong criteria | ||
| BS1 |
|
≥ 0.1% among gnomAD controls, using the subpopulation with the highest frequency and at least 1000 allele counts |
| BS2 | Observed in the homozygous state in a healthy adult | ≥ 3 counts: BS2_strong; ≥ 2 counts: BS2_supp |
| BS3 | Well-established in vitro or in vivo functional studies shows no damaging effect on protein function | BMPR2 functional assay documentation with appropriate controls |
| BS4 | Lack of segregation in affected members of a family | Lack of disease segregation among variant heterozygotes should not be scored due to low penetrance of PAH variants in families. Lack of variant segregation in affected family members should be scored |
| Supporting criteria | ||
| BP1 | Missense variant in gene where only loss-of-function causes disease | N/A |
| BP2 | Observed in trans with a pathogenic variant for a fully penetrant dominant gene/disorder; or observed in cis with a pathogenic variant in any inheritance pattern | N/A |
| BP3 | In-frame deletions/insertions in a repetitive region without a known function | — |
| BP4 | Multiple lines of computational evidence suggest no impact on gene or gene product | REVEL ≤ 0.25, no up/downgrading. SpliceAI ≤ 0.1 for noncanonical splice variants. If no REVEL score or SpliceAI prediction is available, then CADD ≤ 10 can be used |
| BP5 | Variant found in a case with an alternate molecular basis for disease | N/A |
| BP6 | Reputable source recently reports variant as benign, but the evidence is not available to the laboratory to perform an independent evaluation | N/A not to be used |
| BP7 | A synonymous (silent) variant for which splicing prediction algorithms predict no impact to the splice consensus sequence nor the creation of a new splice site and the nucleotide is not highly conserved | — |
- Abbreviations: mod, moderate; PAH, pulmonary arterial hypertension; supp, supporting; VUS, variant of uncertain significance.
Similar to PM2, we considered the highest allele frequency among ancestral subgroups on gnomAD control datasets to apply BA1 as a stand-alone criterion for benign variants with an allele frequency > 1% or ≥ 0.1% for BS1, an allele frequency which was greater than expected for PAH in controls. The presence of homozygotes was scored with BS2 as supporting for ≥ 2 controls and as strong for ≥ 3 controls.
3.1.2. Relevant Variant Frequency in Patients: PS4, PP1, and BS4
Various criteria address the variant frequency in related or unrelated H/IPAH patients. While disease-causing BMPR2 variants are often novel, a recurrence in unrelated individuals can be used as evidence for criterion PS4. If a variant has a very low population frequency (PM2, see above) and the same variant has been identified previously in > 4 unrelated H/IPAH patients, PS4_strong can be applied (Table 1). The criterion is moderate for > 3 previously described cases and supporting for > 1. Recurrent variants were identified through ClinVar, together with unpublished data from the labs of VCEP members and a thorough literature search for published variants not deposited in ClinVar. Importantly, care was taken to avoid counting duplicate reports of the same or related individuals across different publications. Also, cosegregation with disease lends strength to a variant. Analogous to the cardiomyopathy working group [17], we used PP1 as strong with ≥ 7 meioses, moderate for ≥ 5 meioses across or within families, and supporting for ≥ 3 meioses within a single family. In contrast, the lack of segregation of a variant with the disease in an affected PAH patient can be used as benign evidence for BS4. Due to the reduced penetrance of BMPR2 variants, their presence in unaffected family members was not considered evidence for benignity.
3.1.3. Null Variant Decision Tree: PVS1
The criterion with the strongest evidence for pathogenicity is PVS1 (Table 1). The general PVS1 decision tree [18] was adapted for BMPR2 (see Supporting Information 1: Figure S1). The criterion applies to predicted null variants leading to a loss of function. Thus, it mainly refers to nonsense variants introducing a premature stop codon or frameshift variants and canonical splice site variants (+1, +2, −1, and −2) likely inducing nonsense-mediated decay (NMD). For such variants affecting BMPR2 Exons 1 to 11 and all but the final 50 nucleotides of penultimate Exon 12 (c.1_c.2816), “PVS1_very strong” could be applied. A scoring as “strong” was possible if splice site variants were located in the canonical splice donor or acceptor site (positions +1, +2, −1, or −2) and were predicted to result in exon skipping, leading to an in-frame transcript with a loss or truncation of the ligand domain p.33_131, the transmembrane domain p.151_171, the kinase domain p.203_504, or the heterodimerization domain p.485_492.
3.1.4. Functional Assay Characterisation: PS3 and BS3
The PS3 criterion involves in vitro or in vivo functional studies supportive of a damaging effect on the BMPR2 gene or protein according to the ACMG guidelines. To determine whether a variant met the PS3 criterion, we considered quantitative and qualitative assays, as described below. To be considered valid evidence, quantitative assays needed to include biological and technical replicates, positive controls (wild-type BMPR2/BMPR2 and endogenous or total protein), validation controls (previously established, known pathogenic or benign variants within the same assay), negative controls (empty vectors), and robust statistical analysis (usually t-test or ANOVA). Assessment of functional effects did not need to be binary; statistically supported partial effects were also considered. Qualitative cytoplasmic retention assays did not require statistical analysis to apply PS3 as long as they included appropriate controls and replicates. Functional studies that failed to include validation controls could apply PS3 at the supporting level [19] at the discretion of the curator and PH VCEP.
3.1.4.1. Gene Reporter/Luciferase Assays
Luciferase assays may be employed to investigate the functional impact of missense variants. Typically, cells are transfected with a combination of plasmids containing (i) BMP-responsive element from a SMAD promoter incorporated with a luciferase gene; (ii) wild-type and mutant BMPR2, for example, introduced by site-directed mutagenesis; (iii) and cognate Type I receptors (ALK1/2/3/6). Luciferase activity is then measured and normalised to beta-galactosidase or alkaline phosphatase activity. When compared to wild-type BMPR2, these analyses either confirmed abrogation of SMAD-mediated signalling activity for previously established kinase-dead variants or identified variants that retained enzymatic activity with a luciferase signal comparable to wild type [20, 21].
3.1.4.2. Cell Proliferation Assays
The gold standard for assessing changes in cell proliferation is to quantify the percentage of DNA-synthesizing cells. This can be accomplished by measuring the incorporation of thymidine, or thymidine analogs bromodeoxyuridine (BrdU) or 5-ethynyl-2 ′-deoxyuridine (EdU), into de novo DNA. In PAH, the proliferation of human pulmonary arterial smooth muscle, endothelial, or microvascular endothelial cells from H/IPAH transplant patients is compared to unaffected controls (i.e., unused donor tissue). The proliferation of similar cell types from mice heterozygous for patient-specific Bmpr2 variants can be compared to wild type [22]. Growth curve proliferation assays involving direct cell counting are often included as an independent assessment of cell proliferation [22, 23]. Basic controls should include full growth medium with 10% fetal bovine serum (FBS) (positive control) and serum-restricted medium with 0.1% FBS (negative).
3.1.4.3. Protein Binding Assays
Protein binding assays are designed to investigate the interaction of a protein with another protein, ligand–receptor binding, or transient protein binding for signal transduction. The acceptable protein assays in our PH VCEP panel were qualitative immunoprecipitation, glutathione S-transferase (GST) pull-down, and quantitative radioligand binding assays [20, 21, 24–28]. We considered confirmed ligands (bone morphogenetic proteins, BMPs) or coreceptors (activin receptors, ALKs) as acceptable binding partners, while unconfirmed ligands or receptors were not scored. Variant location within BMPR2 domains, for example, ligand binding domain or kinase domain, was also taken into consideration during the assessment of protein binding assays [21, 26, 28]. In addition, downstream effector readouts were limited to SMADs and p38, while any unconfirmed molecules were not assessed. Positive control of wild-type BMPR2 and negative control of empty vector were considered for upgrading or downgrading the functional criteria.
3.1.4.4. SMAD Phosphorylation Assays
For SMAD phosphorylation assays, the ability of a BMPR2 variant to phosphorylate SMAD proteins should be compared to controls, preferably unphosphorylated total SMAD proteins or alternatively, housekeeping genes on a western blot with densitometry. Negative controls were either cells without transfected BMPR2, with BMPR2 wild type, or cells from non-PAH controls. Variants that demonstrated no deleterious effect with levels comparable to wild-type BMPR2 were scored under BS3 or BS3_supporting if validation controls were lacking.
3.1.5. Definition of Critical Domains: PM1
For the PM1 criterion, critical BMPR2 functional domains were defined as the extracellular or ligand binding domain and serine–threonine kinase domain, containing a heterodimerization motif, separated by a transmembrane domain. These domains are delineated by strong evidence from structural studies of the activin Type II receptor extracellular domain, evolutionary conservation across eukaryotic kinases, and hydrogen deuterium exchange mass spectrometry analysis of the interface between BMPR2 and the ALK2 kinase domain. Moreover, gene reporter assays have indicated the relative importance of amino acid residues within these regions [20, 21, 29–31]. Ten cysteine residues within the extracellular domain are required for formation of five disulphide bonds essential for correct three-dimensional folding (p.Cys34, p.Cys60, p.Cys66, p.Cys84, p.Cys94, p.Cys99, p.Cys116, p.Cys117, p.Cys118, and p.Cys123). Sequences from 60 aligned eukaryotic kinases indicate the presence of eight evolutionarily invariant residues (p.Gly212, p.Lys230, p.Asp333, p.Asn338, p.Asp351, p.Glu386, p.Asp405, and p.Arg491) and four nearly invariant residues (p.Gly210, p.Glu/Asn245, p.Gly353, and p.Gly410). Moreover, structural analyses demonstrated that BMPR2/ALK heterodimerization is contingent on amino acids p.Asp485_Leu492 [29–31]. Variants located at these critical amino acid residues were upgraded to PM1_strong. The transmembrane domain was considered a critical domain for loss-of-function variants (nonsense/frameshift/splice site) involving Exon 4 but not for missense variants.
3.1.6. Minor Adaptations: PM5 and PS1
In this study, PM5 at the moderate level was applied when a missense variant was found in the same amino acid as a previously described pathogenic missense variant. If the previous missense variant was classified as Likely Pathogenic, then PM5 was applied at the supporting level. PS1 could be applied as strong if the same amino acid substitution was described previously as pathogenic and as moderate if the variant was considered Likely Pathogenic. Alternative variants were curated using our PH VCEP specifications rather than relying on existing interpretations published in ClinVar or elsewhere. Similarly, PS1 may be applied to exonic or intronic variants with predicted splicing effects based on similarity with a previous pathogenic or Likely Pathogenic variant, weighted according to the ClinGen SVI Splicing Subgroup recommendations [32].
3.1.7. In Silico Tools: PP3/BP4/BP7
The PP3 and BP4 criteria consider the computational evidence that supports the presence (PP3) or absence (BP4) of a predicted, deleterious effect on BMPR2. Our VCEP adopted the rare exome variant ensemble learner (REVEL) [33], SpliceAI [34], and combined annotation-dependent depletion (CADD) [35] scores to apply PP3 (REVEL ≥ 0.75; SpliceAI ≥ 0.2) or BP4 (REVEL ≤ 0.25; SpliceAI ≤ 0.1). In cases when REVEL or SpliceAI scores were not available, a CADD ≥ 20 (PP3) or ≤ 10 (BP4) was considered [35, 36]. SpliceAI ≤ 0.1 was used to apply BP7 for both synonymous and deep intronic variants and could be used together with BP4, as recommended by the ClinGen SVI Working Group.
3.1.8. Other Criteria and Overall Scoring
Specifications for all criteria are detailed in Table 1 and Supporting Information 1: Figure S1. The de novo criteria (PS2 and PM6) and deletion/insertion change criteria (PM4 and BP3) were applied unaltered. PM3, PP2, PP4, BP1, BP2, and BP5 were considered not applicable to BMPR2-related PAH. Criteria referring to a “reputable source” (PP5 and BP6) are no longer used according to ClinGen revised guidance [37]. Criteria were combined using the Bayesian points scoring system outlined by Tavtigian et al. [38] with a modification for likely benign (LB), as follows: Pathogenic (P), ≥ 10 points; Likely Pathogenic, 6 to 9; variant of uncertain significance (VUS), −1 to 5; Likely Benign, −2 to −6; and Benign (B), ≤ −7.
3.2. Pilot Testing of PH VCEP Specifications
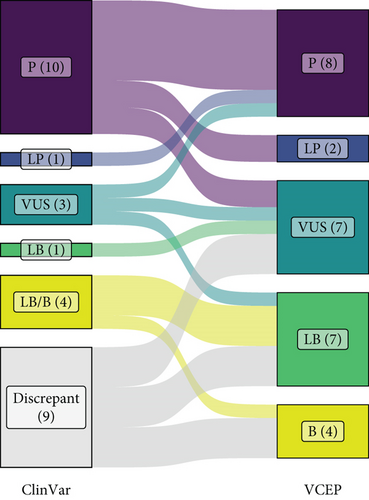
The results of our pilot curation of 28 BMPR2 variants are detailed in Table 2 and Figure 2. The classification of seven variants (25%; six pathogenic and one VUS) did not change from the existing ClinVar classification. These included well-established Pathogenic variants that were included as “positive controls” to test our specifications. If the P/LP and B/LB classifications are grouped per their clinical utility, a further seven variants were unchanged, including four with mixed B/LB classifications in ClinVar. Three of these were resolved to Likely Benign and one to Benign when our specifications were applied, although this is not consequential for their clinical interpretation. Of the remaining 14 variants (50%), nine had discrepant classifications in ClinVar. Eight of these were VUS/LB or VUS/LB/B, of which three were resolved to Benign, three to Likely Benign, and two remained as VUS. Two variants changed from Likely Benign to VUS or vice versa. The most pronounced and clinically consequential changes were seen with intronic splice region variants, with three variants (c.247+1_247+7del, c.529+2T>C, and c.968-3C>G) reclassified from pathogenic or P/VUS to VUS and one from VUS to Pathogenic (c.968-5A>G). As discussed further below, mRNA analysis to confirm the predicted consequences of splice site variants is critical for accurate classification, especially for variants outside of the canonical positions.
| Varianta | Variant type | ClinVar classification | PH VCEP classification | ACMG/AMP criteria and points applied | Scoreb |
|---|---|---|---|---|---|
| c.-924A>G | 5 ′-Untranslated region | VUS/likely benign | Benign | BA1 −8, BS2_supp −1, BP4 −1 | −10 |
| c.-669G>A | 5 ′-Untranslated region | Likely benign/benign | Benign | BA1 −8 | −8 |
| c.218C>G (p.Ser73 ∗) | Nonsense | Pathogenic | Pathogenic | PVS1 +8, PS4_supp +1, PM2_supp +1 | +10 |
| c.240_241insT (p.Lys81 ∗) | Nonsense | Pathogenic | Likely Pathogenic | PVS1 +8, PM2_supp +1 | +9 |
| c.247+1_247+7del | Splice region/intronic | Pathogenic | VUS | PVS1_strong +4, PM2_supp +1 | +5 |
| c.251G>A (p.Cys84Tyr) | Missense | Pathogenic | Pathogenic | PS4_mod +2, PM1_strong +4, PM2_supp +1, PM5 +2, PP3 +1 | +10 |
| c.354T>G (p.Cys118Trp) | Missense | Pathogenic (3) | Pathogenic | PS3 +4, PS4 +4, PM1_strong +4, PM2_supp +1, PM5_supp +1, PP1 +1, PP3 +1 | +16 |
| c.419-38del | Splice region/intronic | VUS/likely benign/benign | Benign | BA1 −8, BS2 −4, BP4 −1, BP7 −1 | −14 |
| c.529+2T>C | Canonical splice site | Pathogenic | VUS | PVS1_strong +4, PM2_supp +1 | +5 |
| c.545G>A (p.Gly182Asp) | Missense | VUS (3)/likely benign (1) | Likely benign | BS3 −4, PP3 +1 | −3 |
| c.797G>C (p.Arg266Thr) | Missense | VUS | VUS | PM2_supp +1 | +1 |
| c.901T>C (p.Ser301Pro) | Missense | Likely pathogenic | Pathogenic | PS3 +4, PS4 +4, PM1 +2, PM2_supp +1, PM6 +2 | +13 |
| c.968-5A>G | Splice site/intronic | VUS (3) | Pathogenic | PVS1 (RNA) +8, PS4_supp +1, PM2_supp +1 | +10 |
| c.968-3C>G | Splice site/intronic | Pathogenic/VUS | VUS | PS4_supp +1, PM2_supp +1, PP3 +1 | +3 |
| c.1040G>A (p.Cys347Tyr) | Missense | Pathogenic | Likely pathogenic | PS3_supp +1, PS4 +4, PM2_supp +1, PM5_supp +1, PP3 +1 | +8 |
| c.1042G>A (p.Val348Ile) | Missense | Likely benign/benign | Likely benign | BS1 −4, PS3_supp +1 | −3 |
| c.1413+1G>A | Canonical splice site | Pathogenic | Pathogenic | PVS1 +8, PS4_mod +2, PM2_supp +1 | +11 |
| c.1424C>A (p.Ser475 ∗) | Nonsense | Pathogenic (2) | Pathogenic | PVS1 +8, PS4_supp +1, PM2_supp +1 | +10 |
| c.1472G>A (p.Arg491Gln) | Missense | Pathogenic | Pathogenic | PS2 +4, PS3_supp +1, PS4 +4, PM1_strong +4, PM2_supp +1, PM5 +2, PP1 +1, PP3 +1 | +18 |
| c.1481C>T (p.Ala494Val) | Missense | Likely benign | VUS | PM1 +2, PP3 +1, BS1 −4 | −1 |
| c.1509A>C (p.Glu503Asp) | Missense | VUS | Likely benign | BS3 −4 | −4 |
| c.1698T>A (p.Ile566=) | Synonymous | VUS/likely benign | VUS | PM2_supp +1, BP4 −1, BP7 −1 | −1 |
| c.1766A>G (p.Tyr589Cys) | Missense | Likely benign/benign | Likely benign | BS1 −4 | −4 |
| c.2186G>C (p.Gly729Ala) | Missense | VUS/likely benign | Likely benign | BS1 −4 | −4 |
| c.2352C>T (p.Val784=) | Synonymous | VUS/likely benign | VUS | PM2_supp +1, BP4 −1, BP7 −1 | −1 |
| c.2618G>A (p.Arg873Gln) | Missense | VUS/likely benign | Benign | BS1 −4, BS3_supp −1, BS4 −4 | −9 |
| c.2887G>T (p.Gly963Cys) | Missense | Likely benign/benign | Likely benign | BS1 −4 | −4 |
| c.2948G>A (p.Arg983Gln) | Missense | VUS (3)/likely benign (1)/benign (1) | Likely benign | BS1 −4 | −4 |
- Abbreviations: mod, moderate; supp, supporting; VUS, variant of uncertain significance.
- aVariant nomenclature refers to transcript NM_001204.7.
- bScoring modified from Tavtigian et al. [38]: pathogenic (P), ≥ 10 points; likely pathogenic (LP), 6 to 9; VUS, −1 to 5; likely benign (LB), −2 to −6; benign (B), ≤ −7.
3.3. Evaluation of Newer Bioinformatics Tools: AlphaMissense and BayesDel
Lastly, we performed a pilot evaluation of two newer bioinformatic prediction tools not covered in the original ACMG/AMP guidelines. As a novel and emerging in silico prediction tool for missense variants, AlphaMissense, developed by Google DeepMind, calculates missense variant pathogenicity by combining structural context and evolutionary conservation, using data from AlphaFold prediction and human and primate variant frequency databases [39]. It generates scores between 0 and 1, with higher scores predicting a greater likelihood of pathogenicity. Suggested thresholds for Likely Pathogenic and Likely Benign are > 0.564 and < 0.340, respectively. BayesDel (no AF) is a deleteriousness metascore. The range of the score is from −1.29334 to 0.75731. The higher the score, the more likely the variant is pathogenic [40]. Both programs were applied, in addition to REVEL and CADD, to the 15 missense variants in our pilot trial and compared to our final VCEP classification. In this preliminary analysis, AlphaMissense outperformed REVEL, CADD, and BayesDel by reaching an agreement with our experts for 12/15 variants, while the other three programs each only supported 5/15 assessments (Table 3).
| Varianta | PHVCEP classification | REVEL | CADD | AlphaMissense | BayesDel |
|---|---|---|---|---|---|
| c.251G>A (p.Cys84Tyr) | Pathogenic | Deleterious (0.95) | Deleterious (32) | Likely pathogenic (0.998) | Deleterious (strong) (0.59) |
| c.354T>G (p.Cys118Trp) | Pathogenic | Deleterious (0.93) | Deleterious (28.5) | Likely pathogenic(0.999) | Deleterious (strong) (0.53) |
| c.545G>A (p.Gly182Asp) | Likely benign | Deleterious (0.81) | Deleterious (28.0) | Uncertain (0.425) | Deleterious (moderate) (0.36) |
| c.797G>C (p.Arg266Thr) | VUS | Uncertain (0.63) | Deleterious (29.5) | Likely pathogenic(0.893) | Deleterious (supporting) (0.21) |
| c.901T>C (p.Ser301Pro) | Pathogenic | Uncertain (0.45) | Deleterious (25.1) | Likely pathogenic(0.905) | Deleterious (supporting) (0.16) |
| c.1040G>A (p.Cys347Tyr) | Likely pathogenic | Deleterious (0.94) | Deleterious (31) | Likely pathogenic (0.994) | Deleterious (moderate) (0.48) |
| c.1042G>A (p.Val348Ile) | Likely benign | Uncertain (0.74) | Deleterious (26.2) | Likely benign(0.201) | Deleterious (moderate) (0.39) |
| c.1472G>A (p.Arg491Gln) | Pathogenic | Deleterious (0.96) | Deleterious (34) | Likely pathogenic(0.992) | Deleterious (strong) (0.62) |
| c.1481C>T (p.Ala494Val) | VUS | Deleterious (0.87) | Deleterious (31) | Likely pathogenic(0.979) | Deleterious (moderate) (0.4) |
| c.1509A>C (p.Glu503Asp) | Likely benign | Uncertain (0.66) | Uncertain (17.7) | Likely benign (0.26) | Deleterious (moderate) (0.29) |
| c.1766A>G (p.Tyr589Cys) | Likely benign | Uncertain (0.58) | Deleterious (28) | Likely benign (0.263) | Deleterious (supporting) (0.18) |
| c.2186G>C (p.Gly729Ala) | Likely benign | Uncertain (0.36) | Deleterious (23.6) | Likely benign (0.101) | Uncertain (0.06) |
| c.2618G>A (p.Arg873Gln) | Benign | Uncertain (0.55) | Deleterious (26) | Likely benign (0.266) | Deleterious (moderate) (0.34) |
| c.2887G>T (p.Gly963Cys) | Likely benign | Uncertain (0.42) | Deleterious (26.8) | Likely benign (0.099) | Uncertain (−0.04) |
| c.2948G>A (p.Arg983Gln) | Likely benign | Uncertain (0.4) | Deleterious (24.8) | Likely benign (0.321) | Uncertain (−0.04) |
| Agreement with VCEP | — | 5/15 | 5/15 | 12/15 | 5/15 |
- Note: AlphaMissense: likely pathogenic, 0.564–1.0; uncertain, 0.34–0.564; and likely benign, 0–0.34. BayesDel: ≥ 0.5, strong pathogenic evidence; 0.27–0.5, moderate pathogenic; 0.13–0.27, supporting pathogenic; −0.36 to −0.18, supporting benign; and score ≤ −0.36, moderate benign [40]. CADD: ≥ 20 pathogenic, 10–20 uncertain, and ≤ 10 benign. REVEL: ≥ 0.75 pathogenic, 0.25–0.75 uncertain, and ≤ 0.25 benign.
- Abbreviation: VUS, variant of uncertain significance.
- aVariant nomenclature refers to transcript NM_001204.7.
4. Discussion
We adapted the general ACMG/AMP variant specification criteria specifically to BMPR2, the gene most commonly implicated in PAH, with loss or reduced function as the mechanism of action. This guidance should be valuable to improve harmonization in reporting variants identified in patients with heritable or idiopathic PAH across clinical laboratories. Of note, we did not consider variants identified in patients with associated PAH; therefore, these specifications may require modification in such cases.
Unlike null variants such as canonical splice site or frameshift variants, it is often challenging to classify novel and rare missense variants as Likely Pathogenic/Pathogenic with currently available data. In many cases, functional data are lacking, and previous reports of the same variant in other PAH patients are usually limited. This often leaves only PM2 (low population frequency) and PP3 (pathogenicity predictions) to support missense variant interpretation. Hence, it is crucial for patients and clinicians that all variants are uploaded to ClinVar to potentially identify recurrent variants or amino acid changes to apply additional criteria (PS4, PS1, and PM5). Accurate predictive testing for the familial disease-causing variant is only possible for Likely Pathogenic or Pathogenic variants, and genetic risk prediction is critical as there are increasingly effective therapies for PAH that are likely more effective when applied earlier in the disease course.
Of the 28 BMPR2 variants curated in this pilot study, only eight (28.6%) had functional analyses that could be scored, of which five provided evidence for PS3, two for BS3, and one for BS3_supp. Access to tissues, cell lines, and nucleic acids to perform the functional analyses considered in this study is not always possible, and, when available, the materials and resources may be limited. The lack of functional analyses presents challenges when determining the impact of a given reported genomic variant. In this regard, the assessment of mRNA in patients with suspected splice site variants located more than two base pairs within the intron is relatively straightforward and provides a key experimental indication of pathogenicity. The importance of this is highlighted by three splice site variants in our pilot panel, including one at the +2 position of a splice donor site, that were downgraded from Pathogenic to VUS due to the lack of supporting mRNA data. Conversely, one variant in the −5 splice acceptor position was upgraded from VUS to Pathogenic on the basis of mRNA analysis, performed in the lab of a VCEP member, that provided evidence for PVS1 (RNA).
Our pilot testing of AI-based prediction algorithms, including AlphaMissense, suggests that such programs may be highly accurate in predicting the pathogenicity of missense variants in the absence of functional data. However, this is an evolving field, with ongoing development of new programs and calibration of threshold values for established tools. Of note, while this manuscript was in revision, a new posterior probability-based calibration was proposed for AlphaMissense [41]. The minimal threshold to apply PP3 was determined as ≥ 0.792, considerably higher than the original 0.564, while the threshold for BP4 moved from 0.340 to < 0.170. As a result, the agreement with our VCEP classification for BMPR2 missense variants was reduced to 7 of 15 variants, with five previously concordant variants changing from Likely Benign to uncertain (Supporting Information 2: Table S1). Nonetheless, Bergquist et al. emphasize that these tools should not be considered the sole classifier; rather, an integrated analysis per the ACMG framework will almost always be needed [41]. With this in mind, we reframed our comparison of these algorithms with our integrated VCEP curation to focus on whether the score produced by each program was concordant, neutral, or discordant with the final VCEP classification, rather than applying a definitive in silico classification. Uncertain calls were considered neutral. The updated thresholds rendered AlphaMissense more conservative in calling evidence for benignity (BP4), with only 2/8 Benign/Likely Benign variants achieving BP4 and the remaining six having indeterminate scores. Importantly, all Pathogenic/Likely Pathogenic variants still met the minimal threshold for PP3, and no AlphaMissense calls were discordant (Supporting Information 2: Table S1). In contrast, CADD scores, even when recalibrated to the more stringent thresholds proposed by Pejaver et al. [40], were discordant for five variants, while the recalibrated thresholds for REVEL and BayesDel [41] were discordant in three and five cases, respectively. While these results suggest that AlphaMissense and REVEL are promising tools, the use of in silico prediction programs is a dynamic landscape that will require ongoing reevaluation. Therefore, the composite approach adopted in our BMPR2 guidelines remains the most robust method of determining variant pathogenicity or benignity [40, 41].
In this study, we encountered a large Iberian family in which 32 individuals were genotyped and 22 were heterozygous for c.1472G>A p.Arg491Gln cosegregating with PAH [42]. However, segregation analyses are typically challenging due to small family sizes, the work needed to organize family studies, and incomplete penetrance. Due to the need for cardiac catheterization for clinical diagnosis, pulmonary hemodynamics may not be available for some family members. In addition, without enough affected heterozygotes, the number of meioses observed may not be sufficient to use the segregation criteria PP1 or BS4. The presence or absence of this evidence can significantly influence the final classification of a given variant. Therefore, it is essential to develop protocols or policies to collect relevant data from large families when there is suspicion of HPAH. Moreover, clinical guidelines recommend offering cascade testing to potentially identify at-risk heterozygotes and offer clinical screening to diagnose PAH as early as possible [5].
ACMG guidelines were developed for Mendelian genetic disorders with the goal of dichotomizing variants into Pathogenic/Likely Pathogenic and Benign/Likely Benign categories to define their clinical utility (or lack thereof). Most heritable PAH cases, including those related to BMPR2, show autosomal dominant inheritance, and thus, our VCEP appropriately sought to adapt ACMG guidelines for this gene. However, PAH shows reduced penetrance, suggesting that other genetic and/or environmental modifiers also play a role. In this regard, the publication by Masson et al. is thought provoking, recognizing that some VUS may demonstrate disease relevance based on population genetics or functional data but do not satisfy the stringent criteria for Pathogenic or Likely Pathogenic classification [43]. Thus, they proposed new terminology of “predisposing” and “likely predisposing.” This concept may be helpful in classifying lower penetrance variants that could contribute to PAH in a polygenic model and should be considered in the future.
In conclusion, these newly adapted ClinGen specification guidelines to assess BMPR2 variants for PAH patients will allow more reliable and accurate variant classification. Although the overall proportion of VUS increased, this reflects that approximately one-third of the chosen variants had discrepant classifications in ClinVar. Notably, we were able to resolve six of the eight variants with a discrepant VUS/LB, reclassifying them as Benign or Likely Benign. The remaining VUS may be reclassified with increasing evidence in the future. Benign or Likely Benign variants are most likely unrelated to the disease and thus do not increase the genetic risk of developing PAH. In contrast, Pathogenic or Likely Pathogenic variants allow for risk stratification of other family members. Thus, the consensus specification document substantially aids in providing patients and caregivers with an accurate genetic diagnosis which may play an increasing role in therapy.
Ethics Statement
All data were obtained from ClinVar and/or the published literature and were fully deidentified. Therefore, IRB approval and informed consent was not required.
Disclosure
This manuscript was submitted as a preprint at the following link [44]: https://www.medrxiv.org/content/10.1101/2024.11.24.24317862v1
Conflicts of Interest
CAE received honoraria for lectures and presentations from OMT and MSD, consulting fees from MSD. CAE is coinventor of the issued European patent “Gene panel specific for pulmonary hypertension and its uses” (EP3507380). CAE works at a laboratory that performs fee for service testing in genes that have been specified by the ClinGen PH VCEP. FC works at a French healthcare hospital (Pitié-Salpêtrière Hospital in Paris) laboratory which performs noncommercial fee for service testing in BMPR2 and other PAH genes. DD works at a Dutch healthcare hospital laboratory which performs noncommercial fee for service testing in BMPR2 and other PAH genes. ME worked at a French healthcare hospital laboratory which performs noncommercial fee for service testing in BMPR2 and other PAH genes. DM works for GeneDx, a laboratory that performs fee for service genetic testing. WKC serves on the Board of Directors of Prime Medicine. GMV and SB were graduate students at the time the study was performed but are now employed by fee for service genetic testing companies, Tempus and Baylor Genetics, respectively. MAA, SB, KMD, and GMV received salary support from the National Institutes of Health. All other authors declare no conflict of interest.
Author Contributions
Conceptualization, methodology, and data curation: all authors; supervision: C.L.W. and W.K.C.; writing—original draft: C.A.E., G.M.-V., R.D.M., S.G., M.S., C.L.W., and M.A.A.; writing—review and editing: all authors
Funding
This research is supported by the National Heart Lung and Blood Institute (R35HL140019) (MAA, SB, and KMD) and Diversity Supplement (R35HL140019-S1) (GMV).
Supporting Information
Additional supporting information can be found online in the Supporting Information section.
Open Research
Data Availability Statement
The BMPR2 specifications are available in the ClinGen Criteria Specification Registry (https://cspec.genome.network/cspec/ui/svi/doc/GN125). Variant curations were published to the ClinGen Evidence Repository and submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/?term=bmpr2%255Bgene%255D%26redir=gene).




