ClinVar database of global familial hypercholesterolemia-associated DNA variants
Funding information: The ClinGen consortium is funded by the National Human Genome Research Institute of the National Institutes of Health through the following grants and contracts: U41HG006834, U41HG009649, U41HG009650, U01HG007436, and U01HG007437. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
For the ClinGen/ClinVar Special Issue
Abstract
Accurate and consistent variant classification is imperative for incorporation of rapidly developing sequencing technologies into genomic medicine for improved patient care. An essential requirement for achieving standardized and reliable variant interpretation is data sharing, facilitated by a centralized open-source database. Familial hypercholesterolemia (FH) is an exemplar of the utility of such a resource: it has a high incidence, a favorable prognosis with early intervention and treatment, and cascade screening can be offered to families if a causative variant is identified. ClinVar, an NCBI-funded resource, has become the primary repository for clinically relevant variants in Mendelian disease, including FH. Here, we present the concerted efforts made by the Clinical Genome Resource, through the FH Variant Curation Expert Panel and global FH community, to increase submission of FH-associated variants into ClinVar. Variant-level data was categorized by submitter, variant characteristics, classification method, and available supporting data. To further reform interpretation of FH-associated variants, areas for improvement in variant submissions were identified; these include a need for more detailed submissions and submission of supporting variant-level data, both retrospectively and prospectively. Collaborating to provide thorough, reliable evidence-based variant interpretation will ultimately improve the care of FH patients.
1 INTRODUCTION
Familial hypercholesterolemia (FH) is an autosomal codominant disorder, characterized by elevated low-density lipoprotein (LDL) cholesterol (LDL-C) levels causing premature atherosclerotic cardiovascular disease when left untreated. FH affects an estimated one in 250 individuals worldwide (Akioyamen et al., 2017), and is considered to be the most frequent monogenic disorder encountered in clinical practice. Since the 1970s, a vast number of potentially pathogenic DNA variants have been identified in FH patients, primarily within LDLR (the gene encoding the LDL receptor), and more recently in other genes involved in LDL metabolism: APOB and PCSK9 (genes encoding apolipoprotein B and proprotein convertase subtilisin/kexin type 9, respectively). Characterizing the genetic etiology of FH has improved our understanding of disease pathophysiology and associated risks, in addition to improving patient management (Defesche et al., 2017; Goldstein & Brown, 2009).
Interpreting the clinical significance of genetic variants is challenging (Bland et al., 2018), and has direct implications for diagnosis and family-based “cascade” screening. While improvements in variant interpretation may aid in the implementation of genetic testing into more routine FH clinical care, this is enhanced when there is information on variants from multiple independent sources that can be shared among laboratories. This data-sharing culture is not new among the FH field; for years the Leiden Open Source Variation Database (LOVD) has served as a publicly available FH-variant repository, hosting 1,707 unique LDLR variants as of 2016 (Leigh et al., 2017). However, ClinVar, an NCBI-funded resource, has since emerged as the primary centralized database for archiving clinically relevant variants for many Mendelian diseases, including FH. ClinVar facilitates a comprehensive approach to both the consolidation and presentation of patient and molecular data, and includes a multitude of interconnected resources to aid in improving variant interpretation (Harrison et al., 2016).
Prior to 2016, there were 331 total (278 unique) FH-associated variant submissions in ClinVar. Here, we present the recent efforts made by the Clinical Genome (ClinGen) Resource consortium, along with various global FH researchers to update the number and characterization of FH variants hosted by ClinVar to aid in the accurate knowledge and interpretation of FH variants. Specifically, we break down the number of FH variants now hosted on ClinVar by gene, location, type, and classification; in addition to providing variant-level characterizations. We then discuss the implications learned from these variant-level and aggregate results.
2 METHODS
2.1 ClinGen FH Variant Curation Expert Panel
The ClinGen FH Variant Curation Expert Panel (FH VC-EP) is composed of >20 members (Supporting Information Table S1). Members were selected on the basis of achieving a balanced representation of expert clinicians, clinical laboratory diagnosticians, researchers, and genomic medicine specialists. An emphasis was also placed on global representation, with members from the United States, Brazil, United Kingdom, the Netherlands, France, Portugal, Czech Republic, Spain, Israel, Australia, and Canada. The FH VC-EP is a part of the ClinGen Cardiovascular Clinical Domain Working Group.
2.2 Variant submission to ClinVar
Starting in 2016, several sources were recruited for consolidation of FH-associated variants into ClinVar. These efforts were facilitated by the FH Foundation working with ClinGen leadership to convene a session of interested parties, including members of the FH VC-EP at the 2016 international FH Summit in Dallas, TX, and 2017 in Miami, FL. First, FH VC-EP members began submitting FH-associated variants and variant-level data from their respective internal databases to ClinVar. We then encouraged global colleagues to submit internally stored FH-associated variants, with a focus on the largest remaining sequencing centers from various countries and jurisdictions. Further, we facilitated variant transfer from existing centralized databases, namely LOVD (https://databases.lovd.nl/shared/genes/LDLR).
Submitters followed a standard protocol for submission. They were required to register their organization/center on the ClinVar Submission Portal (https://submit.ncbi.nlm.nih.gov/clinvar/). Following ClinVar approval, variant submissions were performed using the Submission Template spreadsheet (https://www.ncbi.nlm.nih.gov/clinvar/docs/submit/). Submitted variants required standardized annotation (human genome variation society (HGVS) expression or chromosomal coordinate change), associated condition, interpretation of clinical and/or functional significance, interpretation criteria, collection method (clinical testing or research), allele origin (germline or somatic), and individual affected status. A wide range of additional variant-level data types were optional for inclusion, such as number of variant observations, ethnicity and/or geographic origin of the individual, co-segregation/family data, functional data, phenotypic information, and/or normolipidemic screening results.
2.3 ClinVar variant analysis
Following submission efforts, ClinVar Miner (https://clinvarminer.genetics.utah.edu/) was used to extract variant-level data from the ClinVar database for LDLR, APOB, and PCSK9. Variants that did not have a submitted disease association of FH or accepted alternative term were removed manually, specifically: “familial hypobetalipoproteinemia” (n = 221), “hypercholesterolemia, autosomal dominant, type B; hypobetalipoproteinemia, familial, 1” (n = 156; entry of two opposing conditions per single individual), “LDL-C level quantitative trait locus 1” (n = 3), “hypocholesterolemia” (n = 2), “hypobetalipoproteinemia, familial, 1” (n = 2), “early-onset coronary artery disease (CAD)” (n = 2; removed as other dyslipidemias/morbidities can lead to CAD), “hypobetalipoproteinemia” (n = 1), “C0950123: inborn genetic diseases” (n = 1), “not specified” (n = 191), and “not provided” (n = 164). Variant consequences were determined manually from DNA and protein-level variant information and confirmed using the Mutalyzer Name Checker batch tool v.2.0.28 (Leiden University Medical Center, Leiden, the Netherlands; https://mutalyzer.nl/).
3 RESULTS
3.1 Global ClinVar submission
Prior to 2016, there were 242 (193 unique) LDLR, 63 (59) APOB, and 26 (26) PCSK9 variant submissions present in ClinVar. In a concerted effort to increase this number, the ClinGen FH VC-EP encouraged the submission of FH-associated variants by colleagues and sequencing centers on a global scale. As a result, the number of FH-associated submissions now residing in the ClinVar database increased ∼18-fold and is summarized in Table 1. Additionally, there are 201 LDLR, 423 APOB, and 119 PCSK9 variant submissions that do not have a disease association of FH and were removed from analysis. A total of 30 centers from 13 different countries have submitted FH-associated variants to ClinVar. Submitting center totals are listed per gene in Table 2.
| LDLR | APOB | PCSK9 | Total | |
|---|---|---|---|---|
| All variants submitted to ClinVar | 5,174 | 1,003 | 474 | 6,651 |
| Variants detected in FH patients | 4,973 | 580 | 355 | 5,908 |
| Unique variants detected in FH patients | 2,314 | 353 | 216 | 2,883 |
| Submitting centers | Country | LDLR | APOB | PCSK9 | Total |
|---|---|---|---|---|---|
| LDLR-Leiden Open Source Variation Database, British Heart Foundation | United Kingdom | 1,670 | – | – | 1,670 |
| Laboratory of Molecular Diagnostics, Vascular Medicine, Academic Medical Centre, University of Amsterdam | Netherlands | 686 | 25 | 46 | 757 |
| Centre of Molecular Genetics, Obesity and Dyslipidemias Unit, Pitié-Salpêtrière University Hospital | France | 414 | 1 | 19 | 434 |
| Cardiovascular Research Group, National Institute of Health Dr. Ricardo Jorge | Portugal | 276 | 53 | 70 | 399 |
| Blackburn Cardiovascular Genetics Laboratory, Robarts Research Institute | Canada | 202 | 137 | 30 | 369 |
| Clinical Services Laboratory, Illumina | USA | 97 | 180 | 85 | 362 |
| Molecular Medicine of Metabolic Diseases Unit (U4M), University of Lille, Regional Hospital Center | France | 344 | – | – | 344 |
| Spanish Familial Hypercholesterolemia Foundation | Spain | 320 | 10 | 1 | 331 |
| Laboratory of Genetics and Molecular Cardiology, University of São Paulo | Brazil | 201 | 63 | 16 | 280 |
| Molecular Genetics Laboratory, Centre for Cardiovascular Surgery and Transplantation | Czech Republic | 197 | – | – | 197 |
| Invitae | USA | 156 | – | 40 | 196 |
| Cardiovascular Genetics Laboratory, PathWest Laboratory Medicine WA | Australia | 152 | – | – | 152 |
| Color Genomics | USA | 23 | 65 | 25 | 113 |
| Other | USA, Germany, Finland, India, South Korea | 235 | 46 | 23 | 304 |
- Centers that have submitted >100 FH-associated variants are listed, remaining centers are grouped in “Other.”
3.2 FH-associated variant characteristics
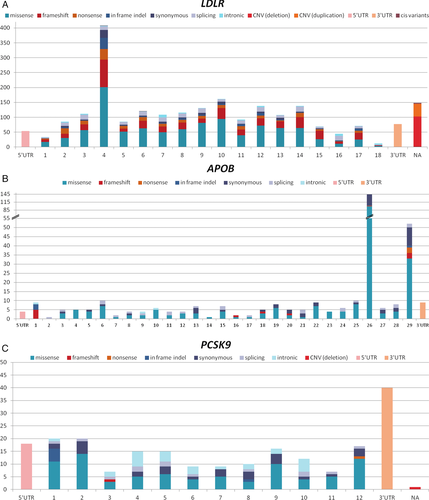
Unique FH-associated variants present on ClinVar are categorized by type for LDLR, APOB, and PCSK9 in Table 3 and shown by location across all exons in Figure 1. Missense variants are the most prevalent unique variant type in each of the three genes, followed by frameshift variants in LDLR, and synonymous variants in both APOB and PCSK9. In LDLR 18% of all unique variants are located in exon 4, in APOB 41% are in exon 26 and 15% in exon 29, and in PCSK9 19% are in the 3’UTR (untranslated region). Figure 2 illustrates the relative proportions of variant classifications for each variant type.
| Variant type | LDLR | APOB | PCSK9 |
|---|---|---|---|
| 3’UTR | 77 | 9 | 40 |
| 5’UTR | 54 | 4 | 18 |
| Frameshift | 430 | 12 | 1 |
| In-Frame indels | 87 | 5 | 6 |
| Intronic | 48 | 3 | 26 |
| Splicing | 198 | 24 | 13 |
| CNV (deletion) | 100 | – | 1 |
| CNV (duplication) | 42 | – | – |
| Missense | 1,011 | 218 | 82 |
| Nonsense | 179 | 4 | 1 |
| Synonymous | 83 | 74 | 28 |
| Cis variants | 5 | – | – |
| Total | 2,314 | 353 | 216 |
- In-frame indels: smaller than one exon; intronic: variants after +/−15 nucleotides (nts) in the intron; splicing: variants known to affect splicing + variants within +/−15 nts in the intron; CNVs: one whole exon or more; cis variants (single submission of two variants on same allele) include: three double missense, one in-frame indel + frameshift, one in-frame indel + missense. CNV, copy number variation; indel, insertion or deletion; UTR, untranslated region.
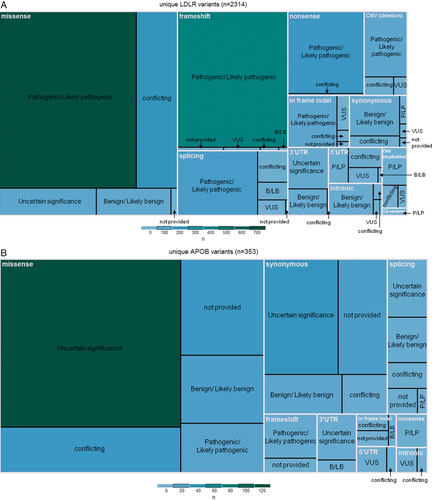
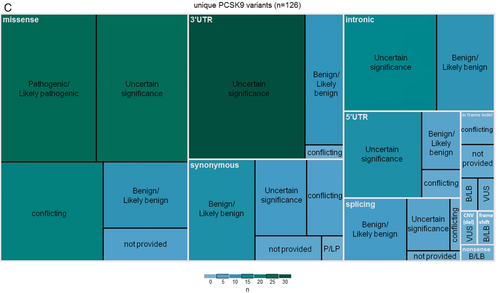
Variants submitted to ClinVar range from benign to pathogenic or can be submitted without an assertion; with the exception of 198 FH-associated variant submissions, submitting centers provided a pathogenicity classification for their variants, found summarized by gene in Table 4. Unique variants are categorized by classification in Table 5; 57.9% (1,670 of 2,883) of these variants have been classified by submitters as pathogenic or likely pathogenic (or both, in cases of multiple submissions for the same variant), 15.5% (448 of 2,883) have been classified as a variant of unknown significance (VUS), and 10.4% (299 of 2,883) have been classified as benign or likely benign. The remaining 13.1% of variants (379 of 2,883) have conflicting classifications using a three-tier system.
| Clinical significance | LDLR | APOB | PCSK9 |
|---|---|---|---|
| Benign | 205 (4.1%) | 57 (9.8%) | 88 (24.8%) |
| Likely benign | 312 (6.3%) | 97 (16.7%) | 54 (15.2%) |
| Uncertain significance | 526 (10.6%) | 254 (43.8%) | 132 (37.2%) |
| Likely pathogenic | 1,525 (30.7%) | 10 (1.7%) | 15 (4.2%) |
| Pathogenic | 2,351 (42.3%) | 42 (7.2%) | 42 (11.8%) |
| Not provided | 54 (1.1%) | 120 (20.7%) | 24 (6.8%) |
| Total | 4,973 | 580 | 355 |
| Clinical significance | LDLR | APOB | PCSK9 |
|---|---|---|---|
| Benign/Likely benign | 200 (8.7%) | 44 (15.1%) | 55 (26.8%) |
| Uncertain significance | 182 (7.9%) | 171 (58.6%) | 95 (46.3%) |
| Pathogenic/Likely pathogenic | 1,614 (70.2%) | 30 (10.3%) | 26 (12.7%) |
| Conflicting classification | 303 (13.2%) | 47 (16.0%) | 29 (14.2%) |
| Not provided | 15 | 61 | 11 |
| Total | 2,314 | 353 | 216 |
- Conflicting classifications (considered for variants with multiple submissions): Benign/Likely benign + Uncertain significance; Pathogenic/Likely pathogenic + Uncertain significance; or Benign/Likely benign + Pathogenic/Likely pathogenic.
3.3 Variant classification methods
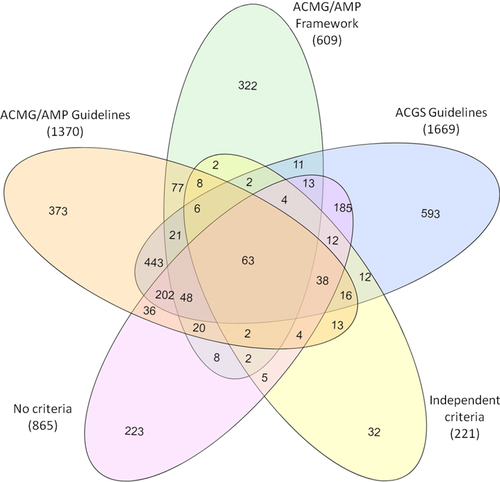
A wide range of criteria has been used to classify FH-associated variants present on ClinVar. These include the general American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) guidelines (2015), specified guidelines adhering to the ACMG/AMP framework, and a number of independent methods. Most variants with multiple submissions have been classified using various different criteria (Figure 3). The specific criteria used by each submitter are listed in Supporting Information Table S2. The most used method was ACMG/AMP framework classification, followed by the Association for Clinical Genetic Science guidelines used in all LOVD-transferred variants. A large number of variants (n = 865) with classifications did not have indication of criteria used (Table 6).
| Criteria used for classification | LDLR | APOB | PCSK9 | Total |
|---|---|---|---|---|
| ACMG/AMP guidelines | 1,144 | 127 | 99 | 1,370 |
| ACMG/AMP framework | 295 | 194 | 120 | 609 |
| ACGS guidelines | 1,669 | – | – | 1,669 |
| Independent methods | 186 | 26 | 9 | 221 |
| No criteria | 793 | 25 | 47 | 865 |
- ACMG/AMP framework: specified criteria adhering to the ACMG/AMP framework; No criteria: classification given but criteria used not provided. ACGS, Association for Clinical Genetic Science; ACMG/AMP, American College of Medical Genetics and Genomics/Association for Molecular Pathology.
3.4 Variant-level data
Some variants (n = 1,972 unique; 3,435 submissions) were submitted with some kind of supporting variant-level data. This included information on patient clinical features, if there was family history of disease, the number of variant alleles or number of families with the variant identified, number of families with observed segregation, if it was an incidental finding and note of any related functional studies published (Table 7). However, information of co-segregation was only submitted to ClinVar for eight variants, and phenotype data was only submitted for 490 unique variants (in 1,043 total submissions). Functional studies were reported for 334 unique variants (437 submissions), the majority submitted as literature review by a single group.
| Variant-level data submitted as evidencea | LDLR | APOB | PCSK9 |
|---|---|---|---|
| Variant alleles/number of families with variant | 1,885 | 26 | 11 |
| Clinical features/Family history | 490 | 0 | 0 |
| Incidental finding | 344 | 0 | 0 |
| Functional study | 293 | 19 | 22 |
| Number of families with observed segregation | 8 | 0 | 0 |
- a Labels extracted directly from ClinVar.
4 DISCUSSION
Data sharing through a centralized open-source database is essential to achieve accurate and consistent interpretation of variants identified during the course of genetic testing. Through the concerted efforts of the ClinGen FH VC-EP, submission of FH-associated variants to ClinVar from different global laboratories resulted in an increase of 10 times the number of unique variants reported during the past years. This was only possible due to a common effort and willingness to share internal data, and demonstrates the power of collaboration across patient groups, academic labs, commercial labs, and scientific funding bodies.
An extensive range of FH-associated variants are now present on ClinVar to aid with variant interpretation. The relative proportions of variants and variant types per gene are consistent with what has been previously reported (Chora, Medeiros, Alves, & Bourbon, 2017; Leigh et al., 2017). However, there are more known FH-associated variants identified in LDLR, APOB, and PCSK9 than previously thought. The FH literature has continued to cite a historical number of ∼2,000 FH-associated variants identified worldwide; however, with ∼2,900 presented here, this has now become outdated.
It is noteworthy that a number of variants with multiple submissions may include instances of “double counting”; a few FH centers have submitted a proportion of their variants to both the LOVD database (in the past) and ClinVar. While the exact number of these variants is presently unknown, efforts are underway to remove such cases. Second, the number of unique copy number variations (CNVs) in LDLR (142; 100 deletions and 42 duplications) may be underestimated quite considerably. There have been 273 total CNV submissions, yet only 12 have defined breakpoints. This is a result of commonly applied detection methods such as multiplex ligation-dependent probe amplification (MLPA) (Wang, Ban, & Hegele, 2005), or more recently next-generation sequencing (NGS) depth of coverage analysis (Iacocca, Wang, et al., 2017), which are limited to exon-level resolution. LDLR CNV submissions in ClinVar have thus largely been grouped by affected exon(s), but the likelihood of each breakpoint being identical in these “unique” CNV types is questionable. Previous breakpoint analysis has shown there are multiple unique CNV events that lead to the deletion of the 5’UTR–exon 1 in LDLR (Hobbs, Leitersdorf, Goldstein, Brown, & Russell, 1988), and the same may be true for other LDLR CNV types.
Only 10.7% of classified variants in LDLR have been considered as VUS by ClinVar submitters, compared to 55.2% and 39.9% VUS in APOB and PCSK9, respectively, suggesting potential pathogenicity is much more difficult to evaluate in APOB/PCSK9 compared to LDLR. Because a loss-of-function in LDLR is a known disease mechanism of FH, any clearly deleterious variant-type in LDLR can be considered pathogenic. However, only very specific variants in APOB and PCSK9 lead to FH. In PCSK9, causative variants must induce a gain of function in the encoded protein, and in APOB, causative variants must allow the production of the protein, but need to specifically alter the binding affinity to LDLR (known LDL-binding domains are located within APOB exons 26 and 29). Generally, any null variant in these genes will lead to hypocholesterolemia, and thus are not expected to be identified in FH patients. This leaves most candidate APOB and PCSK9 variants missense or synonymous, which pose challenges to interpretation. Further, some APOB variants have been shown to have low penetrance, adding another level of difficulty in interpreting variants in this gene (Alves, Etxebarria, Soutar, Martin, & Bourbon, 2014). Accordingly, it is unwarranted to confidently classify variants as pathogenic in APOB and PCSK9 without performing functional studies, leaving many of them as VUS.
This effort has also revealed that many different variant classification methods are being used, which is problematic since non-standardization can lead to different interpretation of identical variants. Indeed, we saw 379 variants (∼15% of variants in each gene) with conflicting classifications. Use of ACMG/AMP guidelines aims to achieve greater standardization and consistency in variant interpretation (Richards et al., 2015). As we saw here, many FH research and diagnostic groups have adopted this new standard. However, the ACMG/AMP guidelines were designed to be generalizable to all Mendelian disorders, and ambiguities leave potential for differences in the application of various criteria among users, yielding inconsistent classifications. For instance, 114 unique variants have conflicting classifications despite all submitters having cited the ACMG/AMP guidelines.
Beyond a degree of inherent subjectivity, the current ACMG/AMP guidelines do not adequately address FH. In a separate study, ACMG/AMP classification of a large subset of FH-associated variants resulted in a large proportion of VUS (42% in LDLR, as well as 90% in APOB and 92% in PCSK9; Chora et al., 2017). Cases of misclassifications when compared against known pathogenic/benign variants were also found. One of ClinGen's key goals is the standardization of gene/disease-specific adjustments to the ACMG/AMP guidelines to address these issues, and to use these specified guidelines to provide a high level of confidence in ClinVar variant classifications. Following a rigorous step-wise process that includes completion of a pilot study and development of a sustained variant curation and discrepancy resolution plan, the FH VC-EP will submit an application for Expert Panel status to the ClinGen Clinical Domain Working Group Oversight Committee for review and final approval. Once approval is obtained, the ClinGen FH VC-EP will curate and classify variants at an Expert Panel review level (3-star, high confidence), with the ultimate goal of reviewing and/or reclassifying all 2,883 unique FH variants on ClinVar using the newly formed FH-specified ACMG/AMP criteria.
Current ClinVar submssions point to specific issues that need to be addressed imminently in order to further improve the interpretation of FH-associated variants, especially in the context of applying FH-specified ACMG/AMP criteria.
First, clinical details accompanying a submission need to have minimum standards. Many LDLR, APOB, and PCSK9 variants were submitted without a disease association, rendering them of little value to curation efforts. Others were submitted with both hyper/hypocholesterolemia associations, and some had potentially incorrect disease associations-–for example, deleterious/null variants in APOB/PCSK9 submitted with a disease association of FH.
Second, richer supporting variant-level data must be submitted. Although FH centers successfully reported numerous variants, the same cannot be said concerning additional supporting variant-level data. Only eight variants had information about co-segregation, and patient phenotype descriptions were nearly nonexistent (e.g., no data on lipid profiles or cardiovascular disease). The large majority of submitters reported no functional studies for detected variants, although this is key to pathogenicity attribution, and are publicly available for more than 300 variants. The ACMG/AMP framework awards points to functional-level data, co-segregation data, normolipidemic data, and the number of observations/unrelated patients with each variant; if this information is kept stored in internal databases it will ultimately have a major negative impact on accurately re-classifying all ClinVar variants. Patient ethnicity would also be useful data, but were unreported.
All submitters should include supporting variant-level data for retrospective and prospective variant submission. Ideally, submissions should include a short summary of phenotype and genetic testing results for each individual, such as untreated LDL-C, the genes tested, and any other variants detected in the patient's sample. As an illustrative case, consider a patient who presents with an LDL-C value typical of heterozygous FH and has a candidate variant in both LDLR and APOB. If the LDLR variant is clearly pathogenic (suggested by previous aggregate evidence) then this case-level information adds evidence to support the APOB variant being benign (if no other evidence is available to suggest otherwise). When two such variants are submitted separately outside the testing context, others might interpret the APOB variant as a VUS or perhaps even as pathogenic if it is the only variant ascertained in their patient and see it has been previously reported on the database. Such contextual interpretation is undoubtedly performed internally by diagnostic laboratories but is currently not part of any variant submission process, despite it being readily accessible at the time of submission.
Third, data submission needs to be ongoing. Although most of the world's largest laboratory repositories for FH variants have now made submissions to ClinVar, a few important populations remain outstanding; including Italy, Denmark, Norway, Germany, Israel, and Japan. Efforts are underway to encourage outstanding centers to submit their variants, and it is imperative this is achieved prior to the reclassification of all variants using FH-specified ACMG/AMP criteria to ensure diverse representation is accounted for in the specification of these criteria. Further, FH-associated variants are likely being identified on an exponential scale as NGS panels are becoming increasingly implemented in routine FH diagnosis (Iacocca & Hegele, 2017), a trend sure to continue as sequencing costs continue to plummet and awareness of FH broadens. Thus, real-time submission of variant data must be a focus for all centers, due to the potential implications this data may have on ACMG/AMP-algorithm-derived variant classifications.
5 CONCLUSION
Efforts of data sharing and reliable variant interpretation are extremely important to improve the care of FH patients. Since FH is so prominent in the population, and as educational efforts continue, more health care/family physicians can be expected to order genetic testing. As such, FH-associated variant submissions to ClinVar are likely to continue to increase. This will also increase the use of ClinVar as an essential resource for variant interpretation, with the goal to reach the largest number of 3-star variants and its corollary in terms of acceleration of the molecular diagnosis of FH, ultimately affecting patient management and cascade screening. The ClinGen FH VC-EP will continue to encourage data sharing and communication between clinical and research FH experts in order to improve variant curation and harmonize FH diagnosis across the world.
ACKNOWLEDGMENTS
The authors would like to thank Illumina Clinical Services Laboratory; JRC acknowledges her PhD fellowship funded by the Science and Technology Foundation (SFRH/BD/108503/2015). TF was supported by the Ministry of Health of the Czech Republic, Grants no. 16–29084A and 15–28277A (all rights reserved). SEH acknowledges funding from the British Heart Foundation for support for the UCL LOVD database (BHF PG08/008) and from the NIHR UCLH BRC.
CONFLICT OF INTEREST
AC has received honoraria from Amgen SAS and Alexion Pharma France SAS. RS has received honoraria related to consulting, lectures, and research activities from Amgen, Astra Zeneca, Akcea, Biolab, Esperion, Kowa, Merck, Novo-Nordisk, Pfizer, and Sanofi/Regeneron. RAH has received honoraria for membership on advisory boards and speakers’ bureaus for Aegerion, Akcea/Ionis, Amgen, Gemphire, and Regeneron/Sanofi. MB received project grants from Sanofi/Regeneron, PRAXIS, and Alexion Pharmaceuticals.