Scaling resolution of variant classification differences in ClinVar between 41 clinical laboratories through an outlier approach
For the ClinGen/ClinVar Special Issue
Abstract
ClinVar provides open access to variant classifications shared from many clinical laboratories. Although most classifications are consistent across laboratories, classification differences exist. To facilitate resolution of classification differences on a large scale, clinical laboratories were encouraged to reassess outlier classifications of variants with medically significant differences (MSDs). Outliers were identified by first comparing ClinVar submissions from 41 clinical laboratories to detect variants with MSDs between the laboratories (650 variants). Next, MSDs were filtered for variants with ≥3 classifications (244 variants), of which 87.6% (213 variants) had a majority consensus in ClinVar, thus allowing for identification of outlier classifications in need of reassessment. Laboratories with outlier classifications were sent a custom report and encouraged to reassess variants. Results were returned for 204 (96%) variants, of which 62.3% (127) were resolved. Of those 127, 64.6% (82) were resolved due to reassessment prompted by this study and 35.4% (45) resolved by a previously completed reassessment. This study demonstrates a scalable approach to classification resolution and capitalizes on the value of data sharing within ClinVar. These activities will help the community move toward more consistent variant classifications, which will improve the care of patients with, or at risk for, genetic disorders.
1 INTRODUCTION
Since its release in 2012, ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) has become an invaluable resource for the genetics community with currently over 583,000 submitted variant classifications from 925 submitters across 64 countries (data accessed March 2018; Landrum et al., 2018). The ClinVar database, maintained by the National Center for Biotechnology Information, archives and aggregates submitted variant classifications and indicates whether they are concordant or discordant within highest review status. Efforts by the National Institutes of Health funded Clinical Genome Resource (ClinGen; https://clinicalgenome.org) to share genetic data have facilitated adoption of data sharing in ClinVar, particularly from clinical laboratories (Rehm et al., 2015). Although most variant classifications from clinical laboratories had previously been unavailable to the genetics community, classifications from clinical laboratories currently account for >83% of all submissions to ClinVar (500,100/600,610; March 2018). Given the range of submitter types, ClinVar assigns each submission a review status and asks submitters to provide additional annotations, such as classification context, collection method, date of variant evaluation, and supporting evidence. These features assist ClinVar users in understanding the context of each classification and what level of review and evidence supports the submitted classification.
Publications assessing the volume of classification differences in ClinVar have produced conflicting findings, often varying by whether annotations such as collection method and review status are accounted for (Dolinsky et al., 2017; Lincoln et al., 2017; Nussbaum, Yang, & Lincoln, 2017) or not accounted for (Balmaña et al., 2016; Gradishar, Johnson, Brown, Mundt, & Manley, 2017). A comprehensive paper analyzing all ClinVar variants classified by two or more submitters (Yang et al., 2017) found 81% concordance when including one- and two-step differences between the three major classification levels (pathogenic [P]/likely pathogenic [LP], variants of uncertain significance [VUS], and likely benign [LB]/benign [B]) and 94.1% concordance when limiting to medically significant differences (MSDs; P/LP vs. VUS/LB/B). When including variants that had a majority consensus, meaning at least two thirds of the classifications were in the same group, concordance between submitters increased to 96.7% (Yang et al., 2017). The difference between complete concordance (94.1%) and majority consensus (96.7%) indicates that of the variants with MSDs between submitters, a subset reach majority consensus with an outlier classification(s) accounting for the discordance. Additionally, when looking at date ranges of outlier submissions, Yang et al. (2017) found that submissions pre-2014 were significantly more likely to be outliers compared to post-2014, suggesting that older submissions are also a significant factor in discordance rates.
Regardless of the true volume of classification differences, sharing variant classifications facilitates identification of differences and allows data sharers the opportunity to reassess and potentially resolve differences. A recent study comparing BRCA1 and BRCA2 variant classifications across 11 clinical laboratories sharing through the Canadian Open Genetics Repository found an initial 26.7% classification discordance rate (Lebo et al., 2018). Upon sharing and comparing classifications, the discordance rate decreased to 14.2%, with the majority of discrepancies resolved by new evidence (44.8%) or revised classification criteria (35.3%). Another study comparing variant interpretations across five hypertrophic cardiomyopathy centers participating in the Sarcomeric Human Cardiomyopathy Registry found an initial 20.5% discordance rate (Furqan et al., 2017). Reassessment and data sharing prompted by the registry reduced the discordance rate to 10.7%, with 35% resolved by review of new data and 13% resolved by reassessment with current classification practices. Both examples illustrate the point that sharing variant classifications and subsequent notification of classification differences can serve as a prompt for reassessment that might not occur within a single laboratory given the rarity of Mendelian disease variants.
In a pilot project from ClinGen's Sequence Variant Inter-Laboratory Discrepancy Resolution group, four clinical laboratories (Ambry Genetics, GeneDx, Partners HealthCare Laboratory for Molecular Medicine, and University of Chicago Genetic Services Laboratories) collaborated to investigate the ability of laboratories to resolve differences through reassessment and data sharing and to identify trends that could educate future plans for discrepancy resolution between all ClinVar submitters (Harrison et al., 2017). Laboratories resolved 87.2% of discordant variants reviewed (211/242), of which 36% were resolved by application of updated classification criteria and 17% were resolved because the reinterpretation had already been completed internally but had not yet been updated in ClinVar.
Although the pilot project was successful in both resolving classification differences and identifying trends in classification differences between clinical laboratories, the pilot only included four clinical laboratories and required a major time commitment (an estimated 1–2 hr per variant per laboratory) as participating laboratories were encouraged to reassess all identified variants with classification differences. To improve efficiency and tackle the variants most likely to be resolved without exchange of detailed datasets, our working group began a second phase of variant classification resolution, which included two modifications in scope compared to the pilot project. First, this working group expanded its membership from 4 to 41 clinical laboratories to help elevate the review status of more variants in ClinVar from a “criteria provided, conflicting interpretations” status to a “criteria provided, multiple submitters, no conflicts” status, which requires that all submitters have concordant classifications. Second, given the increase in volume due to involvement of more clinical laboratories, we first focused on outlier classifications. As results from the pilot project indicated that ∼53% of variants were resolved by reassessment or a recently completed reclassification not yet shared in ClinVar, this focus on outlier classifications on variants with MSDs in ClinVar was anticipated to resolve a large number of discrepancies while minimizing the total time commitment for all laboratories to participate. Although the classifications achieving majority consensus are not necessarily more accurate, this approach was chosen as an efficient means of identifying classification differences that are more likely to be resolved with limited efforts (only one laboratory reassessing) as well as those due to differences in classification algorithms, changes in available evidence, or outdated data in ClinVar.
2 MATERIALS AND METHODS
2.1 ClinVar data analysis
Our analyses used the ClinVar April 2017 submission_summary file (released April 5, 2017; ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/tab_delimited/) to identify all variants in ClinVar with submissions from two or more of the selected 41 clinical laboratories. Using the three major classification levels (P/LP, VUS, and LB/B), classifications for each variant were compared to determine concordance or discordance, and if discordant, the level of discordance was assigned. For variants with greater than two classification terms submitted, the two most discordant terms were used to assign the level of discordance. For example, a variant with a pathogenic, uncertain significance, and benign classifications would be categorized by the pathogenic versus benign difference. To allow for comparison analyses, the same process, but for all ClinVar submitters, was used to determine concordance/discordance parameters for the entire ClinVar dataset.
To identify outlier classifications in ClinVar, variants with MSDs (defined as P/LP vs. VUS/LB/B) were further analyzed to identify a subset that have a majority consensus classification, meaning at least two thirds of submitted classifications are concordant (i.e., two of three, three of four, four of five, four of six, etc.), with one or more outlier classifications. For example, a variant with two submitted likely pathogenic classifications, one submitted pathogenic classification, and one submitted uncertain significance classification would have a pathogenic/likely pathogenic majority consensus (three of four classifications in agreement) and the uncertain significance classification would be categorized as an outlier classification.
2.2 Laboratory data analysis
Each laboratory with at least one outlier classification on a variant with a MSD was sent a custom report (Supporting Information File S1). Laboratories were asked to first determine if the variant had already been reclassified internally but not yet updated in ClinVar, and if not, were asked to reassess the variant with current evidence and methods. If the laboratory's classification was still discordant after reassessment, the laboratory was asked to share the evidence and rationale contributing to the classification. For statistical comparisons, Fisher's exact test was used. A P-value < 0.01 was used as a threshold for statistical significance.
3 RESULTS
3.1 Initial ClinVar concordance data
As of April 2017, the ClinVar dataset included 446,552 submissions from 686 submitters resulting in 294,136 unique variants. In total, there were 49,242 variants with submitted classifications using standard American College of Medical Genetics and Genomics (ACMG)/Association for Molecular Pathology (AMP) terminology from two or more submitters, thus facilitating classification comparison; 77.8% (38,514 variants) had concordant classifications, whereas 17.9% (8,789 variants) had VUS to LB/B conflicting classifications; 3.1% (1,547 variants) had P/LP versus VUS differences and 1.2% (574 variants) had P/LP versus LB/B difference, for a total of 4.3% (2,121 variants) having MSDs (Figure 1).
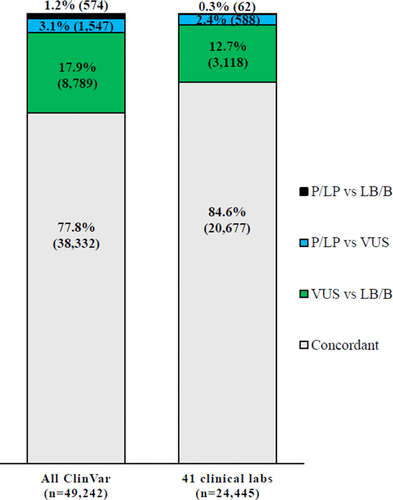
To determine concordance and discordance between clinical laboratory submitters in ClinVar, a set of 41 clinical laboratories were selected from the list of ClinVar submitters based on submitter name and number of variants (Supporting Information Table S1). As of April 2017, these laboratories had submitted 188,766 variant classifications to ClinVar (range per submitter 31–58,383 variant classifications). Classification comparison identified 24,445 variants from 1,351 genes interpreted by at least two of the 41 selected laboratories. Of the 24,445 shared variants, 84.6% (20,677 variants) had concordant classifications between laboratories on the three-tier scale (Figure 1). Only 2.7% (650 variants) of the 24,445 shared variants had MSDs, which are most likely to impact medical management. The majority of MSDs (90.5%; 588 variants) were P/LP versus VUS differences and only 62 variants of this set (9.5%) had P/LP versus LB/B differences. The remaining 12.7% (3118 variants) of all shared variants were VUS versus benign (LB/B) differences, which are less likely to affect medical management, though often lead to differences in counseling and clinical follow-up time.
3.2 Outlier analysis
Six hundred fifty variants with MSDs were further refined to determine which variants had a majority consensus in ClinVar (Figure 2). As the majority consensus determination requires more than two-thirds consensus, only MSDs with three of more submitters were analyzed (244 variants). Of the 244 variants, 87.3% (213 variants) had a majority consensus at the time in ClinVar. Due to low volume of each, variants that either (a) had a LB/B majority consensus (n = 12) or (b) had both VUS and LB/B classifications when combined resulted in a nonpathogenic majority consensus (n = 12; for example, a variant with five submitters: one pathogenic, three VUS, and one benign) were both designated as “VUS/B” majority consensus. Of the 213 variants that had a majority consensus, 47.9% (102 variants) were a pathogenic majority consensus with VUS outlier(s), 40.8% (87 variants) were a VUS majority consensus with P/LP outlier(s), and 11.3% (24 variants) were a VUS/B consensus with P/LP outlier(s) (Figure 2).
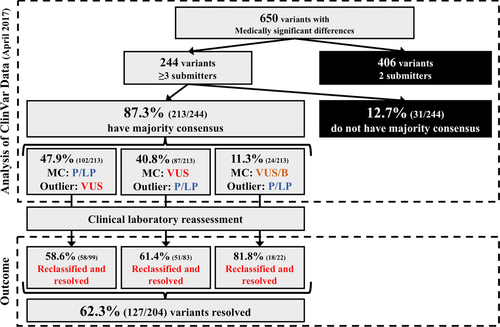
To determine outlier trends, dates of evaluation provided in ClinVar were compared for outlier classifications to classifications contributing to the majority consensus. In total, there were 771 individual classifications for the 213 variants with outlier classifications, of which 96.9% (747/771 classifications) were submitted to ClinVar with an evaluation date provided by the submitter. For the 213 outlier classifications, six classifications were not submitted with an evaluation date and were excluded from analysis of outlier dates. Of the outlier classifications, 36.2% (75 variants) were classified in 2014 or beforehand and 63.8% (132 variants) were classified in 2015 or more recently. When comparing the outlier classification evaluation date to those contributing to the majority consensus, 38.2% (79 variants) of outlier classifications had the oldest evaluation date. In contrast, only 19.8% (41 variants) of outlier classifications had the most recent evaluation date. For the remaining 43.7% (93 variants) of outlier classifications, there were both newer and older classifications contributing to the majority consensus.
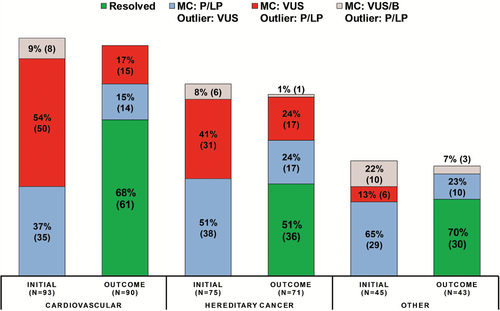
Two hundred thirteen variants with outlier interpretations were from 87 genes; however, 50 genes had only one variant each. Three genes accounted for 25.3% of all variants with an outlier interpretation, MYBPC3 (22 variants), MYH7 (21 variants), and CHEK2 (11 variants), all three of which are genes causative for autosomal dominant conditions with reduced penetrance. To determine if there were outlier trends by disease area, genes were placed into disease categories (Figure 3): cardiovascular (31 genes; 93 variants), hereditary cancer (27 genes; 75 variants), and other (29 genes; 45 variants). However, there were no statistically significant differences in outlier rates across disease areas.
3.3 Laboratory reassessment
Although the initial analysis for concordance included classifications from 41 clinical laboratory ClinVar submitters, only 17 laboratories had one or more outlier classifications (Supporting Information Table S1). Overall, 17 laboratories with one or more outlier classifications had a higher volume of submissions in ClinVar (mean: 9,784 variant classifications; range: 78–58,383) compared to the 24 laboratories with no outlier classifications (mean: 934 variant classifications; range: 31–15,738). Additionally, 94.1% (16/17) of laboratories with outlier classifications submitted variants spanning all three major classification levels, whereas only 50% (12/24) of laboratories with no outlier interpretations submitted variants spanning all three major classification levels.
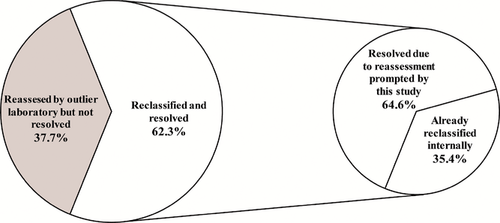
Each of the 17 laboratories with an outlier classification was sent a custom report of outlier classifications and was asked to indicate if the variant had already been reclassified internally or if reassessment was performed upon report of the outlier classification (Supporting Information File S1). Results were not returned for nine variants and those variants are not included in resolved or not resolved counts. Of the 204 variants with results returned, 62.3% (127 variants) were resolved. Of the 127 variants, 64.6% (82 variants) were resolved by reclassification based on reassessment prompted by this project and 35.4% (45 variants) were resolved by a previously completed reassessment but had not yet been updated in ClinVar (Figure 4; Supporting Information Table S2).
Comparing outlier scenarios, there was no statistically significant differences in the resolution rate between any of the three scenarios: majority consensus P/LP outlier VUS variants (58.6%; 58/99 variants), majority consensus VUS outlier P/LP variants (61.4%; 51/83 variants), and majority consensus VUS/B outlier P/LP variants (81.8%; 18/22 variants). However, if focusing only on resolution of the subset of majority consensus VUS/B outlier P/LP variants that were majority consensus LB/B outlier P/LP variants (100% resolution; 11/11 variants), the differences in resolution compared to majority consensus P/LP outlier VUS variants (58.6%; 58/99 variants) were statistically significant (P = 0.0064). Differences in the resolution rate between majority consensus LB/B outlier P/LP variants (100% resolution; 11/11 variants) and majority consensus VUS outlier P/LP variants (61.4%; 51/83 variants) approached but did not meet statistical significance (P = 0.0140). Additionally, there were no significant differences in the resolution rates of outlier types by disease area (Figure 3). However, there was a significant difference in resolution rates comparing variants when the outlier submitter last evaluated the variant in 2014 or older (74.6%; 56/75 variants) compared to when the outlier submitter last evaluated the variant in 2015 or more recently (51.5%; 68/132 variants; P = 0.0016).
For the 77 unresolved variants, evidence and notes returned by the outlier laboratory were analyzed to determine trends for persistent differences. Overall, differences in classification for variants indicated as risk or low-penetrant pathogenic accounted for 7.8% of unresolved variants (six variants total; four majority consensus VUS/B with P/LP outlier variants; two majority consensus VUS with P/LP outlier variants). Additionally, differences in classifying allelic or haplotype variants accounted for 5.2% of unresolved variants (four variants total; three majority consensus P/LP with VUS outlier variants; one majority consensus VUS with P/LP outlier variant).
Lastly, to assess the impact of outlier notification, we compared resolution rates of variants prompted for reassessment by this study to the resolution rate for variants not prompted for reassessment. To assemble this comparison group, we identified variants with MSDs that did not reach a majority consensus in April 2017 (437 variants; Figure 2) and therefore were not included in any reports to individual laboratories. April 2018 ClinVar data were used to determine whether MSDs remained over the time frame of the study. After 1 year, 24.3% (106) of variants that previously had MSDs, but did not have majority consensus in ClinVar and were not reported to individual laboratories as part of this study, became concordant between clinical laboratories in ClinVar. This is a significantly lower proportion (106/437; P < 0.0001) than variants resolved when individual laboratories were prompted to reassess outlier variants (127/204 variants). However, given that the variants that were not prompted for reassessment also did not have a majority consensus, other factors may be contributing to the lower resolution rates.
4 DISCUSSION
Clinical laboratory reassessment, prompted by notification of outlier classifications on variants with MSDs in ClinVar, resulted in 62.3% (127 variants) resolution. Importantly, this resolution rate does not include data sharing between laboratories, which in our pilot project impacted 33% of resolved variants. Instead, the goal of this project was to determine if engagement with clinical laboratories submitting to ClinVar and notification of outlier classifications compared to the majority consensus could resolve interpretation differences in a more time-efficient manner than having all laboratories involved in a discrepancy re-evaluate variants from the start. This approach allows us to first resolve classification differences that are simply due to missing certain sources of evidence identified by subsequent submissions, often not available during an earlier assessment, which minimizes the time commitment from participating laboratories. The fact that only 62.3% of variants were resolved by reassessment by the outlier laboratory suggests that laboratories were not reclassifying outlier classifications just to be in concordance with other clinical laboratories.
As participating laboratories were only queried regarding timing of reclassifications (prior to this study versus the result of a reassessment prompted by this study) and not the rationale for reclassifications, a limitation of this study is that we are unable to determine what evidence types and classification algorithm changes are most likely to impact reclassifications, factors evaluated in our prior study (Harrison et al., 2017). However, we can determine that assessment date is a major reclassification factor as we identified a significantly higher resolution rate when the outlier's previous interpretation was from 2014 or older (74.6%; 56/75 variants) compared to the resolution rate when the outlier's previous interpretation was from 2015 or newer (51.5%; 68/132 variants; P = 0.0016). Although new evidence including the emergence of large-scale population databases such as ExAC (Lek et al., 2016) and gnomAD presumably impacted the higher resolution rate in variants interpreted before 2015, this also correlates with release of the 2015 ACMG and AMP guidelines for variant interpretation (Richards et al. 2015), which may also play a significant role in resolution efforts by promoting consistent interpretations among laboratories. This finding is supported by a 2017 survey of 65 laboratories, which found that 97% of laboratories use the evidence criteria described in the 2015 ACMG/AMP guidelines for variant classification (Niehaus_ACMGAbstract# 258).
General policies for variant reassessment vary by clinical laboratory. However, most laboratories reassess variants when they are observed in an additional case, at the request of providers, and/or with the release of new interpretation guidelines. In our study, 35.4% (45 variants) of resolved variants resulted from internal reclassifications that had not yet been updated in ClinVar, which suggests that for a subset of discrepancies, routine reassessment based on continued observation of the variant will resolve differences. The remaining 64.6% (82 variants) of resolved outlier variants were due to laboratory reassessment prompted by this study. Although a subset of these discrepancies could be resolved over time with additional observations and routine reassessment, many variants identified in Mendelian disease testing are extremely rare, as evidenced by a study showing that 77% of pathogenic, likely pathogenic, and uncertain significance variants from a single laboratory were only observed once (Rehm et al., 2015). Therefore, sharing variant interpretations in ClinVar is critical as an additional ongoing quality assurance measure for laboratory reassessment of rare variants.
Because only the outlier laboratory was prompted to reassess each variant and share their classification rationale and evidence, comparisons to the rationale and evidence used by other laboratories are not available; however, based on data from the outlier laboratories, we can identify variant characteristics likely to result in discordance. For example, these data highlight the need for community consensus around nonstandard variant classification terminology. Indeed, our variants with P versus VUS/B classification differences were unresolved after outlier reassessment because the outlier submitter internally classifies the variant as a risk allele but submitted the variant to ClinVar as benign for a Mendelian disorder. Additionally, of the 18 majority consensus VUS/B outlier P/LP variants resolved, three variants were resolved by a submitter changing their classification to “risk” in ClinVar, as nonstandard classification terms do not factor into concordance in ClinVar. This issue with terminology regarding risk alleles was also noted by Yang et al. (2017), who found that variants that deemed to be “low penetrance” or “risk” by text analysis were less likely to be concordant (49.2%) than observed across all ClinVar data (89.3%), indicating that risk and/or low-penetrance pathogenic variants contribute a significantly disproportionate fraction of discordance in ClinVar. Comparing interpretations across five clinical laboratories, Garber et al. (2016) found only two of 293 variants had pathogenic versus benign interpretation differences, and both were variants that are not disease causing but impact newborn screening results and thus have clinical relevance for the tested individual and provider (Garber et al., 2016). These examples suggest that although P versus B discrepancies are rare (0.3% of all variants classified by ≥2 of the 41 submitters in this study; 62/24,445 variants) and critical to resolve, a subset of these discrepancies may be due to nonstandard variant terminology issues. Thus, the community and ClinVar would benefit from guidance regarding terminology and criteria needed to interpret variants in the spectrum of risk/susceptibility or reduced penetrance as well as clinically relevant variants that are not disease causing.
Results for this study also suggest that differences in how laboratories store and submit allelic variants (such as haplotype and cis/trans variants) to ClinVar can be a cause of classification discordance. Of the total resolved variants (127), two variants that had majority consensus P/LP with VUS outlier were resolved by the outlier laboratory deciding to remove classifications for the individual variants and instead submit a single pathogenic classification for the two variants in cis as these variants have only been identified in cis. However, clinical laboratories contributing to majority consensus each submitted the variants independently as pathogenic with at least one submitter including summary evidence that noted the variant was always found in cis with another variant. Additionally, two variants remained unresolved due to haplotype issues, but for opposite reasons: (a) an outlier submitter retained their pathogenic classification as the variant does cause disease when in cis with a promoter variant and (b) an outlier submitter retained their VUS interpretation as the variant has not been reported independently of a pathogenic variant in cis. These examples indicate that differences in how laboratories structure their classifications when submitting to ClinVar with regard to allelic variants are also a source of discordance in ClinVar. To address the inability to separate the effects of variants in cis on the pathogenicity of an allele when a single variant contributing to pathogenicity cannot be delineated, we suggest that laboratories submit both individual and cis submissions of the variants and submit the same clinical significance classification for all three SCVs (submission to ClinVar: ClinVar assigned accession number for each individual submission). Each SCV should refer to the other SCVs and clearly note that the classification is dependent on the cis state. The two individual SCVs should note that pathogenicity may be altered if the variant is observed without the cis variant.
Variants prompted for resolution in our study had significantly higher resolution rates compared to variants with MSDs that did not reach a majority consensus, and thus were not prompted for reassessment by this study (24.3%; 106/437). We believe that notification of outlier classifications is a successful approach to scaling resolution efforts between clinical laboratories, while minimizing the time commitment from all laboratories. However, given that 67.2% (437/650 variants) of all MSDs between laboratories did not have a majority consensus initially, the outlier approach is not sufficient to address all discordant classifications.
In the next phase of discrepancy resolution, in addition to sharing internal data on unresolved outlier variants, we will also include resolution efforts on MSDs with only two clinical submitters, as these variants accounted for 92.9% (406/437 variants) of all MSDs that did not have a majority consensus. Discrepancies between only two clinical laboratory submitters will be prioritized by identifying pairs of clinical laboratories with a high rate of discordance. By working one-on-one, a pair of laboratories may identify trends in classification algorithm differences between the labs that could facilitate continued resolution and prevent future discrepancies (Garber et al., 2016). In addition, given that date of evaluation was a statistically significant factor in resolution rates, we will request that the laboratory with the older classification evaluate the variant first to minimize the time required by each laboratory. Finally, we will also ask the small number of laboratories that have not publicly shared their evidence for variant classification to reassess their variants first.
The 41 submitters selected for this study do not represent all clinical laboratories sharing data in ClinVar. Future rounds of variant resolution using the majority consensus and outlier approaches will include data from all ClinVar submitters that designate submissions with “clinical testing” as the collection method (227 submitters; April 2018). This task will be facilitated by recent automation of the report generation process, which means that comprehensive reports including all differences, not just outliers, can be sent to clinical laboratories on a routine basis. Laboratories may also identify discrepant classifications through a monthly report of conflicting classifications released by ClinVar (ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/tab_delimited/). This file reports pairwise conflicts in classification, meaning concordance across multiple submitters is difficult to assess. Further, this report is not limited to classifications submitted by clinical laboratories, as it includes classifications submissions by research laboratories and those based solely claims in the literature. These types of submissions are more likely to be outlier classifications compared to clinical testing classifications (Yang et al., 2017). Additionally, new tools have been developed for ClinVar submitters and users to identify custom sets of conflicting classification, such as by submitter or by level of conflict (Butler et al 2018; Henrie et al., 2018).
This study highlights the importance of timeliness of updated classifications to ClinVar. Among the variants resolved in this study, 35.4% resulted from previously completed internal reassessments and reclassifications that had not yet been updated in ClinVar. Although the scope and frequency of ClinVar submissions varies by clinical laboratory, prioritizing resubmission of reclassified variants would help ClinVar be more representative of a laboratory's current classifications. As submission to ClinVar is a manual process, a ClinVar submission interface that allows laboratories to selectively update their previous classifications and evidence in ClinVar without generating a full ClinVar submission form may also help laboratories to update their data more frequently. In addition to more frequent submissions to ClinVar, providing an evaluation date for variant classifications allows ClinVar users to assess how up-to-date classifications are. Of the 771 individual classifications on the 213 variants with MSDs, evaluation dates were provided for 96.9% (747/771 classifications), thus allowing ClinVar users to determine how recently the variant was assessed by each laboratory.
In conclusion, sharing variant classifications in ClinVar allows for comparison between clinical laboratories and identification of variants most in need of reassessment. Although a subset of classification differences will be resolved over time by routine reassessment, notification of outlier classifications can serve as a high-priority prompt for reassessment. Continued discussions with the community regarding terminology for risk variants and how to deposit allelic variants into ClinVar will help increase concordance. Given that the classification of variants for their role in disease requires expert opinion and subjective review of scientific evidence and medical data, complete concordance is not expected. Variant classifications in ClinVar often represent a snapshot in time from the submitting clinical laboratory. Continuous ongoing data sharing, discrepancy resolution, and updating ClinVar will help the community move toward more consistent variant classifications, which will improve the care of patients with, or at risk for, genetic disorders.
CONFLICTS OF INTEREST
All authors are clinical service providers and are employed by laboratories that offer fee-based clinical sequencing. This employment is noted in the author affiliations. The authors declare no additional conflicts of interest beyond their employment affiliation. This publication was supported by the National Human Genome Research Institute of the National Institutes of Health through the U41HG006834 (Rehm) grant. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.