On the verge of diagnosis: Detection, reporting, and investigation of de novo variants in novel genes identified by clinical sequencing
For the ClinGen/ClinVar Special Issue
Abstract
The variable evidence supporting gene–disease associations contributes to the difficulty of accurate variant reporting in a clinical setting. An evidence-based scoring system for evaluating the clinical validity of gene–disease associations, proposed by ClinGen, considers experimental as well as genetic evidence. De novo variants are heavily weighted, given the overall rarity in the genome and their contribution to human disease, however they are reported as “genes of unknown significance” in our center when there is insufficient evidence for the gene–disease assertion. We report a collection of 21 de novo variants in genes of unknown clinical significance ascertained via clinical testing, of which eight of 21 (38%) are predicted to cause loss of function. These genes were subjected to ClinGen scoring to assess the strength of gene–disease relationships. Using a cutoff for moderate high or strong, 10 of 21 genes now have sufficient evidence to qualify as likely pathogenic or pathogenic variants. Sharing such cases with phenotypic data is imperative to strengthen available genetic evidence to ultimately upgrade clinical validity classifications and facilitate accurate molecular diagnosis.
1 INTRODUCTION
Next-generation sequencing (NGS) technologies have revolutionized clinical genetics, with whole exome sequencing or whole genome sequencing (WES/WGS) facilitating diagnoses for patients with genetic disorders, often resulting in changes in the medical management (Lionel et al., 2017; Soden et al., 2014; Vrijenhoek et al., 2015; Willig et al., 2015). Several studies using trio WES/WGS have shown de novo variants, in particular those predicted to be loss of function (LoF), to be a major cause of severe early-onset genetic disorders such as intellectual disability (ID), autism spectrum disorder, and other neurodevelopmental diseases (Goldmann et al., 2016; Kong et al., 2012; Samocha et al., 2014; Wilfert, Sulovari, Turner, Coe, & Eichler, 2017). Such variants are predominantly of paternal origin and increase with advanced paternal age (Goldmann et al., 2016; Kong et al., 2012). De novo variants occur throughout the genome, from single-nucleotide variants (SNVs) to small insertions–deletions (indels) and potentially larger structural variations (Goldmann et al., 2016; Kong et al., 2012; Samocha et al., 2014; Wilfert et al., 2017). The magnitude of the contribution of de novo damaging alterations affecting important genes in development and causing human diseases is still being investigated; however, they are overall rare events, with one to two events expected in coding genes of each generation (Wilfert et al., 2017). Given their rarity in the genome and contribution to genetic disease, de novo variants are heavily weighted in variant classification and should be carefully considered in clinical analysis of WES/WGS.
A rare de novo variant in a dominant gene fitting a patient's phenotype is interpreted as likely pathogenic or, if the variant is LoF, pathogenic. However, interpretive challenges arise when such variants, particularly LoF and/or de novo, are identified in genes that lack sufficient evidence for association with human disease. Although such variants may be compelling, variant interpretation criteria do not apply for genes of unknown significance (GUS). In our center, they are reported as variants of unknown significance, in theory, allowing clinicians to monitor the literature for additional reports. Gene–disease assertions should ideally be confirmed by reports in multiple affected family members and/or occurrence in unrelated individuals with a similar phenotype, which can be problematic for rare Mendelian diseases. As such, it is important to publish case level data to establish new gene–disease assertions with adequate validity for accurate clinical interpretation.
New disease–gene assertions are being made at a stunning rate, with around 40 new and 450 updated entries in OMIM per month (https://omim.org/statistics/update). Assessing the evidence behind such assertions is important to facilitate accurate interpretation of genomic data. New guidelines for vetting such gene–disease assertions have been proposed by the ClinGen working group, which developed a scoring method for gene curation. This method weighs genetic and experimental data in the scientific literature with expert review to classify gene–disease pairs into one of six categories (definitive, strong, moderate, limited, disputed, or refuted) in a semiquantitative manner. At this time, 1,372 of the 4,865 OMIM genes (human phenotype for which the molecular basis is known or presumed) have been curated by ClinGen working groups (https://www.clinicalgenome.org/). A particular need lies in genes associated with ID and autism spectrum disorders (ASD), because many lack OMIM accession numbers. Indeed, many genes associated with neurodevelopmental disorders (NDD) are published in studies with large cohorts with limited information on phenotype and inheritance patterns. Efforts to organize, consolidate, and curate this information to define the clinical relevance of genes and variants are viewable in online databases such as ClinGen and SFARI. In addition, SFARI Gene curates genes associated with ASD in the literature, including both rare and common variants, using a manual multistep review process by an expert panel of researchers. SFARI scores are regularly updated based on publication of new scientific data and feedback from the ASD research community. In-depth annotation of 1,134 rare variants and 12 common variants was recently completed, with 81 new references added, bringing the total number of genes curated in SFARI to ∼990 (https://www.sfari.org/resource/sfari-gene/) (Larson, Arrand, Tantam, Jones, & Holland, 2018; Zhang & Shen, 2017). Of note, less than 50% of SFARI curated genes have an OMIM entry, of which 10% (n = 84) score as high confidence or strong associations.
This study aims to provide case level data on de novo variants in 21 genes reported in patients undergoing clinical WES or WGS. In addition, we examine the evidence for the respective gene–disease associations using ClinGen guidelines for curation. This study highlights the need for sharing case level data needed for gene curation, which ultimately increases the diagnostic utility of NGS in individuals with rare diseases. Such efforts require close collaboration between ordering physicians, molecular geneticists, gene experts, and researchers.
2 MATERIALS AND METHODS
2.1 NGS sequencing, variant calling, and analysis
Nine hundred seventy-one patient samples were referred by subspecialist pediatricians for trio clinical NGS (WES or WGS) with targeted phenotype-driven analysis in a 23-month period. Patients’ clinical records and previous testing results were reviewed prior to testing. Peripheral blood samples were provided for the proband and both parents, when available. DNA was extracted using a Chemagen (Perkin Elmer, MA) following standard procedures. NGS was prepared using the TruSeq PCR free library prep kit, with the addition of 5 cycles of PCR (Illumina, San Diego, CA). Sequencing was completed on an Illumina HiSeq 2500 or 4000 instrument (Illumina, San Diego, CA) utilizing paired end 2 × 125 base pair reads. Samples were sequenced for a mean coverage of ∼35 × (WGS) or ∼80 × (WES). Base calling was performed and required a minimum of 500,000 raw cluster density with 75% passing filter and 80% or greater of reads above Q30. If these quality checks (QC) metrics were satisfied, samples were processed through an alignment and variant detection pipeline using DRAGEN 2.0.4-2.1.3 (Miller et al. 2015); although some older samples were processed using BWA 0.7.2 and GATK 3.2-2. Postpipelining QC included a minimum of 85% of reads aligning to the human genome and a minimum of 85 Gb (WGS) and 6 Gb (WES) of data obtained after alignment is complete. Variant annotation and categorization was performed using Rapid Understanding of Nucleotide variant Effect Software (RUNES v.3.4.3–v4.2.4) as previously described (Saunders et al., 2012; Soden et al., 2014; Willig et al., 2015). Variants were filtered to a 1% minor allele frequency and prioritized by type using VIKING software, as previously described (Saunders et al., 2012; Soden et al., 2014; Willig et al., 2015), and using the American College of Medical Genetics and Genomics guidelines (Richards et al., 2015). Candidate gene lists were generated by SSAGA and/or Phenomizer using Human Phenotype Ontology (Kohler et al., 2014) terms with a cutoff at P value of 0.5. These gene lists were imported into VIKING to guide the analysis, however phenotype and OMIM filters were removed when necessary. Pathogenic, likely pathogenic, and variants of unknown significance in genes related to phenotype were reported; likely benign and benign variants were not reported. For WGS, incidental findings in the 59 genes recommended by ACMG (Kalia et al., 2017) were analyzed if requested by the family, with variant reporting limited to pathogenic and likely pathogenic. No specific CNV caller was used, however manual inspection of alignments was performed as needed. De novo variants (with parental sampled identity confirmed) were reported in GUS. These clinically reported GUS were limited to those with previously published links to a relevant human phenotype. Each GUS was submitted to GeneMatcher (https://www.genematcher.org) (Sobreira, Schiettecatte, Valle, & Hamosh, 2015). De novo variants in genes with no previous reported human phenotype were not reported. This study was approved by the CMH institutional review board.
2.2 Scoresheet for assessing clinical validity of 21 clinically reported GUSs with de novo variants
Gene–disease relationships were scored independently by two PhD molecular geneticists based on ClinGen curation categories: limited (1–6), moderate (7–11), strong (12–18), or definitive (>12 with replicative studies) published. The ClinGen Variant Curation Interface is available for public use (https://github.com/ClinGen/clincoded/wiki/GCI-Curation-Help). The category “definitive” was not applicable in this study because it is generally reserved for Mendelian disease genes that have been reported in independent studies over a period of at least 3 years (Strande et al., 2017). Points were assigned for the number of previously reported, unrelated patients with compelling variants in the respective gene, entries in Deciphering Developmental Disorders (Firth et al., 2009), SFARI (https://www.sfari.org/resource/sfari-gene/) (Larson et al., 2018; Zhang & Shen, 2017), or in large cohort exome or genome studies for NDD. Assessing case-level and case–control data was highly dependent on the inheritance pattern and phenotype reported. As all variants investigated in our cohort were de novo, the number of points for genes associated with autosomal recessive disorders was capped. Second, evidence was downgraded if the report contained different phenotypic findings or variant type inconsistent with the assumed disease mechanism (i.e., a missense variant in a gene associated only with LoF). In addition to genetic evidence, points were given for experimental evidence, gene function, cellular or model organism, protein interactions, pathway, and constraint metrics. The total number of points were used to determine the clinical validity score for each curator. The sum of points were weighted against any available contradictory evidences such as “negative” functional/animal studies, reduced penetrance, and lack of constraint in population databases (such as the residual variation intolerance score [RVIS], gene damage index [GDI], and ExAC constraint scores). Scores assigned by the two independent curators were averaged to determine the final classifications for the 21 gene–disease relationships.
ClinGen scores were compared to SFARI Gene scoring, if available. Briefly, SFARI scores assess the strength of evidence linking candidate genes to ASD (https://www.sfari.org/resource/sfari-gene/), considering genotypes observed in ASD cohorts, functional studies and animal models, and expert opinion/curation from the ASD scientific community. Gene–disease relationships fall into seven possible categories: S (syndromic, genes predisposing to autism in the context of a syndromic disorder such as Fragile X), category 1 (high confidence with genome-wide statistical significance between cases and controls, with independent replication), category 2 (strong: statistical significance between ASD cases and controls), category 3 (suggestive: relatively small studies of candidate genes, using either common or rare variant approaches), category 4 (minimal evidence: relatively small studies of candidate genes with accessory evidence), category 5 (hypothesized but untested: genes that have been implicated solely by evidence in model organisms or other functional nature), and category 6 (genes that have been tested in a ASD cohort, but the weight of the evidence argues against a role in ASD). SFARI curation relies on expert objectivity and complex assessment in the context of genetic heterogeneity in ASD.
3 RESULTS
3.1 Patient characteristics
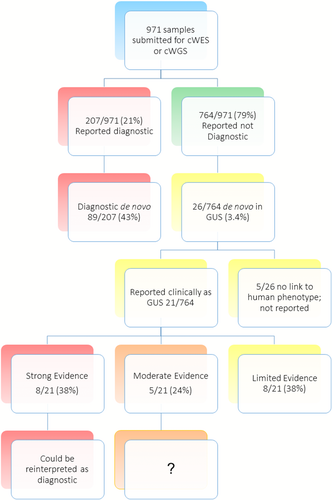
Nine hundred seventy-one patient samples were submitted for clinical NGS in a 23-month period (Figure 1). The overall diagnostic rate was 21% (207/971), with de novo variants accounting for 43% (89/207) of positive cases. An additional 26 (3.4%) patients had heterozygous de novo variants in GUS. Five of these were excluded for lack of a published human phenotype and were not clinically reported (Figure 1). In total, 21 de novo variants in GUS were clinically reported in 20 patients, including ATG5, BPTF, CUL3, EEF2, GCC2, KDM3B, KDM5B, KIAA0100, KMT2C, MYADML, NLGN4Y, PDE10A, REST, RORA, RYR3, SBNO2, TBLIXR1, TRIP4, UBE2H, VPS4A, and ZNF666. Eight of 21 (38%) are predicted to cause LoF (Figure 1 and Table 2). The average number of variants reported in these patients was 10 (Table 1). One patient also had a pathogenic variant in PKD2, consistent with a diagnosis of Polycystic kidney disease; six patients were heterozygous carriers for one (4/6) or two (1/6) autosomal recessive diseases (Table 1). The diagnostic odyssey prior to NGS included multiple blood and urine samples, often involving cerebrospinal fluid punctures, comparative genomic hybridization (CGH) array, multiplex ligation-dependent probe amplification (MLPA®), other NGS panel testing, and biochemical testing (Supporting Information Table S1). The mean age at testing was 6 years (ranging from 3 months to 18 years) with a male/female ratio of 2:1 (Table 1, Supporting Information Table S2). Etiologic testing was performed for a range of clinical concerns and by a variety of subspecialists. The majority of clinical NGS reports (57%) were ordered by geneticists, followed by neurologists (24%), 9% from perinatalogists, and one from gastroenterology (Supporting Information Table S2). A significant number of patients with de novo variants in GUS (95%, 19/20) had NDD, consistent with previous reports of high de novo variant rates in this group of disorders (Table 1, Supporting Information Table S1).
| Family | Sex | Age at analysis | Patient phenotype | Gene | Inheritance pattern | Zygosity | Coding | Protein | Other | Total number of VUS reported |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 1 yrs | Abn pituitary, bilateral CL/CP, deafness, dysphagia, polydactyly, tetralogy of Fallot | EEF2 | AD | het. | c.1784C > T | p.S595F | 7 | |
| 2 | M | 3 yrs | Absent thumb, speech delays, FTT, microcephaly | CUL3 | AD | het. | c.173A > G | p.Y58C | 4 | |
| 3 | M | 7 yrs | FTT, hypotonia, GDD, CL/CP, dysmorphism, deafness, Abn optic nerve | KIAA0100 | AD | het. | c.5954T > C | p.L1985P | 8 | |
| 3 | M | TBL1XR1 | AD | het. | c.1336T > G | p.Y446D | ||||
| 4 | M | 2 yrs | GDD, ptosis, craniosynostosis, lactic acidosis, epileptic spasms, focal T2 hyperintensity | ATG5 | AR | het. | c.62C > G | p.T21R | 8 | |
| 5 | M | 4 yrs | Anxiety, ASD, impulsivity, ID, obesity | VPS4A | AD | het. | c.719_722del | p.Q240Vfs*106 | 10 | |
| 6 | M | 3 yrs | Dolichocephaly, dysphagia, GED, G-tube feeding, hypotonia, GDD, joint laxity | NLGN4Y | X-linked | hemiz. | c.260G > A | p.R87Q | 7 | |
| 7 | F | 4 yrs | Dysmorphia, brain malformation, hypoplasia of optic nerve, hypothyroidism, hypotonia, GDD, heterotopia, hydrocephalus, obesity, seizures, SOD | TRIP4 | AR | het. | c.350C > T | p.A117V | 17 | |
| 8 | M | 4 yrs | Dysmorphia, dysphagia, flat cornea, GDD, hypoplasia of teeth, nystagmus | RYR3 | AD | het. | c.13787C > T | p.S4596F | ORC4, c.1054+1G > T | 12 |
| 9 | M | 6 yrs | Dysmorphia, bruising susceptibility, GDD, FTT, microcephaly | BPTF | AD | het. | c.5660del | p.P1887Rfs*14 | PLA2G6, c.2370_2371del (p.Y790*) | 13 |
| 10 | M | 18 yrs | Dysmorphia, ADHD, ASD, FTT, GDD, seizures | KMT2C | AD | het. | c.7826_7829dup | p. P2611Tfs*8 | ABCA3, c.875A > T (p.E292V) | 6 |
| 11 | M | 8 yrs | Abn of the kidney, ASD, hyperpigmented/hypopigmented macules, GDD, PFO | MYADML2 | AD | het. | c.880G > T | p.N294Y | 12 | |
| 12 | M | 4 yrs | ASD, dysmorphia, hypotonia, GDD, sleep disturbance, strabismus | KDM5B | AD | het. | c.1625_1626del | p.F542Cfs*23 | KIAA0586, c.428del (p.Arg143Lysfs*4); TBCK, c.1031+1G > A | 6 |
| 13 | M | 10 mo | Dysmorphia, hypotonia, FTT, PFO, sleep apnea | REST | AD | het. | c.2464del | p.E822Kfs*21 | 10 | |
| 14 | M | 15 mo | GDD, hydronephrosis, infantile spasms, multiple renal cysts, nephronophthisis | SBNO2 | AD | het. | c.3177_3189dup | p.Y1064Afs*6 | CEP290,c.4437+1G > A | 13 |
| 15 | F | 5 yrs | Dysmorphia, arthralgia, astigmatism, brachycephaly, speech delay, puffy feet/hands, elbow dislocation, obesity, polyhydramnios, sleep apnea | UBE2H | AD | het. | c.136T > C | p.Y46H | 11 | |
| 16 | F | 5 yrs | Situs inversus, heterotaxy, delayed skeletal maturation, duodenal atresia, FTT | KDM3B | AD | het. | c.4549C > T | p.R1517* | 5 | |
| 17 | F | 6 yrs | CHD, aplasia/hypoplasia of the corpus callosum, GDD, hemiplegia, dysmorphia | PDE10A | AD/AR | het. | c.1091+7A > G | p.? | 10 | |
| 18 | F | 3 mo (dcd) | Skeletal abn, brain malform, dysmorphia, ASD, PDA, hydrocephalus, SOD | GCC2 | AD | het. | c.4045C > G | p.Q1349E | 10 | |
| 19 | M | 13 yrs | Dysmorphia, esotropia, global brain atrophy, GDD, GH def, tremor, macrocephaly, renal cyst, seizures, short stature, strabismus | RORA | AD | het. | c.1333C > T | p.R445* | PKD2 c.2143delC (p.Leu715*); NGLY1 c.1516C > T (p.Arg506*) | 8 |
| 20 | F | 9 yrs | ASD, broad-based gait, ID, dysmorphia, regression, GE reflux, hypotonia, GDD, hyperextensibility, macrocephaly, obesity, tall stature | ZNF668 | AD/AR | het. | c.1009A > G | p.K337E | 17 |
- Abn: abnormal; AD: autosomal dominant; ADHD: attention deficit and hyperactivity disorder; arr: genome-wide microarray; AR: autosomal recessive; ASD: autism spectrum disorder; CDG: congenital disorders of glycosylation; CHD: congenital heart defect; CL/CP: cleft lip/cleft palate; dcd: deceased; Del: deletion; Dup: duplication; F: female; FTT: failure to thrive; GDD: global developmental delays; GED: gastroesophageal defect; GH def: growth hormone deficiency; GUS: gene of unknown clinical significance; het.: heterozygous; hemiz.: hemizygous; ID: intellectual disability; LP: likely-pathogenic; M: male; mo: months; PDA: patent ductus arteriosus; PFO: patent foramen ovale; SOD: septo-optic dysplasia; US: ultrasound; VUS: variant of unknown clinical significance; yrs: years.
- *STOP codon.
3.2 ClinGen scoring
To reassess the potential clinical validity of 21 de novo variants reported in GUS, two PhD molecular geneticists independently evaluated each gene using the ClinGen scoring system for gene–disease assertions (Strande et al., 2017). A comparison of points assigned by the two independent curators was completed and final classifications were determined for each gene (Table 2, Supporting Information Tables S3 and S4). Overall there was agreement between the two sets of scores, however, one minor discrepancy between curator classifications was noted for one gene (ATG5), which was classified at the border of limited versus moderate evidence (6.5:9); in this case, one curator weighted the sum of points to account for the inconsistences in the mode of inheritance reported (autosomal recessive vs. de novo AD). The linkage disequilibrium, functional studies, and knockout and knock-in mouse models supported the role of impaired autophagy in neurodegenerative diseases such as spinocerebellar ataxia but were inconsistent for mode of inheritance and the phenotype reported (global developmental delays, ptosis, craniosynostosis, lactic acidosis, epileptic spasms and focal T2 hyperintensity). The scoring system was influenced by the timing of publication, with significant gaps between independent publications suggesting uncertainty and newer published reports lacking time to refute claims. However, broader NGS-based testing led to more convincing publications as compared to linkage and/or candidate gene sequencing.
| Gene | NGS report [Y] | OMIM | OMIM phenotype | OMIM created or modified [Y] | Orphanet | Additional published phenotype [SFARI or Pubmed] | Publication date of article of interest [pubmed] | GMhits | SFARI score | ClinGen consensus score [points] | GUS as LP or P |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Recent publications | |||||||||||
| KIAA0100 | 2015 | ASD | 2012: 22495306 | - | Limited [5] | ||||||
| TBL1XR1 | 2015 | 616944, 602342 | ID, Pierpont Syn. | 2017 | 520; 487825 | ASD, West, SZ | 2012: 23160955; 2016: 26740553, 27479843, 26769062; 2017: 28574232, 28348241, 28687524, 28771251, 28191889, 28588275 | - | Strong | Strong [14] | √ |
| ATG5 | 2016 | 617584 | SCA | 2017 | Ataxia, ID and NDD | 2016: 26812546 | 2 | Moderate [7.5] | |||
| NLGN4Y | 2016 | ASD | 2008: 18628683 | 2 | Minimal | Limited [4] | |||||
| TRIP4 | 2016 | 617066 | MD | 2016 | 486815; 486811 | SMA | 2016: 26924529, 27008887 | 1 | Moderate [11.5]a | ||
| RYR3 | 2016 | ASD, EEE, SZ | 2017: 27513193 | >10 | Moderate [11.5]a | ||||||
| KMT2C | 2016 | 617768 | Kleefstra Syn. | 2017 | 261652 | ASD, ID, NDD, SZ | 2017: 29069077 | 0 | Strong | Strong [15] | √ |
| MYADML2 | 2017 | SZ | 2014: 24463507 | 0 | Limited [4] | ||||||
| REST | 2017 | 617626 | Fibromatosis | 2017 | 2024; 654 | ASD | 2012: 22495311 | 4 | Moderate [10.5]a | ||
| SBNO2 | 2017 | SZ | 2011: 22373040 | 9 | Limited [2] | ||||||
| UBE2H | 2017 | ASD | 2003: 14639049; 2017:28540026 | 2 | Minimal | Limited [5] | |||||
| PDE10A | 2017 | 616921, 616922 | Dyskinesia | 2006, 2017 | 494541; 494526 | NDD, Chorea | 2016: 27058446, 27058447; 2017: 29130591 | 3 | Strong [18] | ||
| GCC2 | 2017 | 2017: 28097321 | 3 | Limited [4] | |||||||
| ZNF668 | 2017 | ASD, dysmorphia with NDD, and FTT | 2012: 22865819; 2016: 26633546 | 0 | Limited [6] | ||||||
| Collaboration on going | |||||||||||
| EEF2 | 2016 | 609306 | SCA | 2014 | 101112 | 5 | Moderate [8] | ||||
| BPTF | 2016 | 617755 | NDD with MCA | 2017 | ID, NDD, MCA | 2017: 28942966 | >10 | Strong [15] | √ | ||
| KDM5B | 2017 | 178469 | ASD, ID, NDD, SZ | 2014: 24307393; 2015: 26785492; 2017: 28554332 | - | Strong | Moderate [11.5]a | √ | |||
| KDM3B | 2017 | SZ | 2014: 25420024; (ASHG meeting 2017) PgmNr 349 | >10 | Strong [15] | √ | |||||
| RORA | 2017 | ASD, NDD | 2014: 25363760; 2018: 28708303 | >10 | Minimal | Strong [17] | √ | ||||
| VPS4A | 2016 | ASD | 2014: 25356899 | 1 | Limited [4] | ||||||
| CUL3 | 2016 | 614496 | PHA | 2018 | 300530 | ASD | 2012: 22495306; 2016: 25969726, 27824329, 27841880; 2017: 28191889, 28263302 | 3 | High | Strong [13.5] | √ |
- √: GUS reinterpreted as likely-pathogenic (LP) or pathogenic (P); -: not reported/present; ADHD: attention deficit and hyperactivity disorder; ASD: autism spectrum disorder; GDD: global developmental delays; GM: GeneMatcher; FTT: failure to thrive; ID: intellectual disability; MCA: multiple congenital anomalies; MD: muscular dystrophy; NDD: neurodevelopmental disorders; PHA: pseudohypoaldosteronism; SCA: spinocerebellar ataxia; Syn.: syndrome; SZ: schizophrenia; Y: years.
- ClinGen Classification: Limited (1–6); moderate (7–11); strong (12–18); definitive (§, strong with replicative studies within 3 years).
- SFARI Classification: Syndromic (S); high confidence (1); strong (2); suggestive (3); minimal (4); hypothesized (5).
- a At the limited of two categories.
- §Replicative studies within 3 years.
3.3 Correlation with OMIM, Orphanet, GeneMatcher, and SFARI gene entries
A minority (43%; 9/21) of reported GUSs had OMIM phenotypes (Table 2), most of which (8/9, 89%) had a creation date months or even years following the date of the clinical report (Table 2). Notably, there was significant overlap with Orphanet status (7/9 [78%] OMIM genes versus 8/21 [38%] total GUSs), with only one in OMIM but absent from Orphanet, and the other in Orphanet and not OMIM (Table 2). Consistent with the observation of OMIM phenotype entry date, 13 of 21 (62%) GUS in this study had publications near or after the time of report, 11 of which scored as having moderate or strong evidence. In addition, we compared the number of GeneMatcher hit(s), with ClinGen scores for the 21 GUS.
A positive correlation was observed between high-scoring GUS and number of GeneMatcher hits, with those with > 10 GeneMatcher hits all scoring as strong or moderate high clinical validity. In addition, 76% (16/21) of our GUS had at least one GeneMatcher hit, of which 37% (6/16) resulted in collaborations to functionally assess GUS, further phenotyping and potential publication.
Interestingly, for the seven genes with SFARI entries, there was strong agreement with ClinGen scoring in six (86%) (Table 2). The discrepant gene, RORA, had a minimal SFARI score but strong ClinGen score due to a very recent publication (Guissart et al., 2018). All four genes with SFARI scores of “high” or “strong” scored as “high” or “moderate high” for ClinGen, and two with a “minimal” SFARI ranking had A “limited” ClinGen classification. No disease–gene associations were scored as “definitive” due to lack of replicative studies over 3 years (Table 2, Supporting Information Tables S3 and S4).
3.4 Other metrics
We used three different published metrics to assess if a GUS is subject to strong selection against variation: (a) the RVIS (Petrovski, Wang, Heinzen, Allen, & Goldstein, 2013), (b) the GDI (Itan et al., 2015), and (c) the ExAC constraint metric (Lek et al., 2016). The RVIS predicts if a gene is more intolerant to variation (i.e., likely to be disease causing) (Petrovski et al., 2013). The RVIS was associated with strong or moderate high clinical validity score in nine of 12 (75%) GUS. However, three genes (CUL3, REST, and TBL1XR1) scored as strong or moderate high clinical validity but have a RVIS between ∼40–97%, indicating high tolerance to variation. The GDI, derived from a genome-wide, gene-level metric of the mutational damage in the general population, has been shown to be an efficient gene-level approach for filtering out false positive variant (Itan et al., 2015), thereby serving as a potential indicator of the relative biological indispensability (low GDI) or redundancy (high GDI) of a given human gene. However, little correlation was observed between the GDI and either RVIS or ClinGen scores (Supporting Information Table S3). Therefore, GDI was not helpful in assessing the clinical validity of the GUSs reported in our cohort.
Finally, we compared ExAC gene-level constraint metrics to ClinGen scores obtained for the 21 GUS. For LoF, three classes of genes with respect to tolerance to LoF variation are assumed. For genes with haploinsufficiency as the disease model, the closest the LoF constraint score (pLI) is to 1, the less tolerant the gene is to LoF (Lek et al., 2016). For missense variants, ExAC gene-level constraint metrics provide a Z score for the deviation of observed counts from the expected number, with positive Z scores indicating intolerance to variation (less variants than expected in the population) and negative Z scores indicating tolerance to variation (more variants than expected in the population) (Lek et al., 2016). The 21 clinically reported GUSs with de novo variants had an average pLI score of 0.7 (range: 0–1) and Z score of 2.5 (range: –1.26 to 5.66). Six of eight (75%) GUS reported with LoF are predicted to be highly intolerant to LoF (pLI: 0.91–1) and five of six (83%) were classified as moderate (1) or strong (4). The two remaining GUS with LoF variants, predicted to tolerate LoF (pLI: 0–0.02), had ClinGen scores of either moderate or limited. In the 12 GUS reported with de novo missense variants, 11 of 12 (9%) had an available Z score, and nine of 11 (81%) were predicted intolerant to variation with an average Z score of 3.15 (range: 0.75–5.66). Of those, five of nine (55%) were classified as moderate (2) or strong (3), and four of nine (45%) had limited association with human disease. In agreement with previous published data, genes encoding for protein in core biological processes and pathways have high constraint metrics for both type of variants (Supporting Information Table S3), providing evidence of putative association with severe Mendelian diseases (Lek et al., 2016). Based on ExAC pLI scores, ∼3,230 genes are highly intolerant to LoF, with less than 30% of those yet to be associated with a human phenotype. The remaining genes may represent good candidates for uncharacterized severe Mendelian diseases or incompatibility to life due to dosage sensitivity. Finally, two of 21 (9%) GUS could not be assessed using these metrics. The first, MYADML2, had no ExAC constraint metrics available, and the second, PDE10A, has very high ExAC constraint scores but the variant in question is a (c.1091+7A > G) intronic substitution with unknown impact on protein. Although good correlation is observed between scores generated by algorithms such as RVIS, GDI, and ExAC, they should be used with caution, particularly in a clinical setting.
4 DISCUSSION
The ClinGen scoring system is useful for vetting gene–disease associations. A helpful next step would be developing consensus for what level of evidence is required to call a GUS clinically valid. Our data support the need to reassess gene validity overtime, as the current ClinGen scores were highly influenced by data published following clinical reporting. Less than 45% of reported GUSs have OMIM or Orphanet phenotypes, most of which (∼90%) had an entry creation date months or even years following the date of the NGS clinical report. If a cutoff of 10.5 or higher is made for ClinGen scoring, 11 of 21 GUS (52%) currently have adequate evidence of clinical validity.
A strong correlation was observed between SFARI and ClinGen scores in our cohort. As a matter of interest, less than 10% of the SFARI curated genes (84/990) fall into categories 1 and 2 (high confidence or strong association), and of those, 33% (28/84) are associated with a syndromic form of ASD. Furthermore, only 60% of those with a high/strong SFARI gene association have an OMIM entry, of which 10% are not associated with ASD or NDD in OMIM (Supporting Information Table S5). For SFARI category 3 curated genes (176/990), less than 60% (53/88) have an OMIM phenotype associated with ASD or NDD (Supporting Information Table S5). The extensive effort deployed by both ClinGen working groups and SFARI gene scoring advisory committees may benefit from sharing data and unifying expertise, in particular for ASD/NDD genes. In addition, a strong association between the number of GeneMatcher hits and strength of ClinVar evidence, with genes with more than 10 hits in GeneMatcher falling in the strong or moderate/strong ClinGen category (> 10.5). This attests to GeneMatcher as well as SFARI being essential tools for finding additional patients for case-level evidence, and assessing the evidence behind gene–disease assertions.
Reporting de novo variants in GUS may offer a future diagnosis for some patients without incurring the cost of a total reanalysis, because a significant number of GUS in our cohort graduate to strong or moderate classifications within 12–18 months after testing. As a consequence of reclassification due to new evidence, addendum reports in five patients in this cohort were issued for genes with strong clinical validity (BPTF, CUL3, KMT2C, RORA, and TBL1XR1). As an example, in case 3, two de novo variants were reported in two genes, TBL1XR1 and KIAA0100, both with weak association with ASD at the time of testing (late 2015). In 2016–2017, few reports emerged with de novo missense and frameshift variants and deletions involving TBL1XR1 in patients with ID and autism, but without any of the dysmorphic findings or malformations (Laskowski et al., 2016; Riehmer et al., 2017; Wang et al., 2016). Moreover, a specific missense variant (p.Tyr446Cys) in TBL1XR1 was later associated with Pierpont syndrome in seven unrelated patients (Laskowski et al., 2016; Slavotinek et al., 2017). Interestingly, our patient was heterozygous for a de novo variant affecting the same residue (c.1336T > G, p.Tyr446Asp) and had clinical findings compatible with Pierpont syndrome. However, it is possible that KIAA0100 plays an additional role in this patient's phenotype, as there is a higher burden of de novo genetic events in syndromic children (Levenson, 2016; Tammimies et al., 2015; Wilfert et al., 2017).
Some genes with well-established OMIM entries require different classifications for atypical phenotypes or alternative modes of inheritance caused by different mutational mechanisms. For example, CUL3 has been associated with autosomal dominant Pseudohypoaldosteronism type IIE (PHA) or Gordon's syndrome (OMIM # 614496). However, all ∼16 PHA disease-associated variants are strictly localized within exon 9 coding or splice junctions (Boyden et al., 2012; McCormick et al., 2014). On the contrary, more than 16 rare CUL3 variants, including seven de novo (CNV, LoF and nonsynonymous SNV), located throughout the gene, have been associated with ASD/NDD with or without congenital malformations in more than eight independent publications (C Yuen et al., 2017; Codina-Sola et al., 2015; De Rubeis et al., 2014; Iossifov et al., 2015; Kong et al., 2012; O'Roak, Vives, Fu, et al., 2012; O'Roak, Vives, Girirajan, et al., 2012; Stessman et al., 2017; Wang et al., 2016). Furthermore, CUL3 is part of the recurrent 16p11.2 CNV implicated in multiple neurological phenotypes for which functional studies suggested that the regulation of the KCTD13-CUL3-RhoA pathway is crucial for early embryonic development, regulating brain size and connectivity (Anderica-Romero, Gonzalez-Herrera, Santamaria, & Pedraza-Chaverri, 2013; Chen et al., 2009; Lin et al., 2015). Three CUL3-associated proteins (KEL-8, Gigaxonin and NAC1) are involved in synaptic plasticity, neurofilament/tubulin architecture, and proteolysis machinery for synaptic remodeling (Anderica-Romero et al., 2013). Thus, in such cases, gene clinical validity may be reached without spectrum clear understanding of genotype–phenotype relationships. Therefore, understanding the biological basis of disease-causing mechanisms as well as genotype–phenotype correlations is essential.
With the rapidly evolving knowledge of gene–disease associations, some would argue that reporting GUS has no benefit to the patient because this may result in over interpretation of an uncertain result. GUS may be confirmed, disputed or refuted over time, however, are potentially beneficial because they promote reinterpretation or reanalysis rather than additional testing. The messy reality of NGS data generating findings of unknown clinical significance (VUS as much as GUS) is not going away anytime soon. Conversely, clear delineation of best clinical practices for reporting criteria of VUS and GUS is still lacking. Many challenges will need to be navigated on a case-by-case basis with collaboration and communication between clinicians, clinical laboratories, and patients. If clinicians disclose a GUS result, the question then arises of how this information is being understood and handled by patients. It is necessary to evaluate the strength of the evidence before ascribing a genotype to a patient's phenotype. As such, a GUS should not be used in clinical decision making; and efforts to resolve the classification of GUS blur the lines between research and clinical service. Patient advocacy and project such as MyGene2 (https://mygene2.org/MyGene2/) promote data sharing, including GUS, as families play key roles in finding additional affected individuals, identify potential collaborative groups, supporting research, and promoting awareness.
The thorough evaluation of the clinical validity of a gene–disease association as undertaken in this study is time consuming. For this reason, some clinical laboratories offer panels of well-vetted genes with clear clinical association over WES/WGS for genetically heterogeneous conditions. This results in a more manageable data size and reduces the burden of VUS reporting. Undoubtedly, the incorporation of experts in fundamental research for gene curation committees may provide more objectivity in the interpretation of certain types of evidence, incorporation of unpublished data, and in the evaluation of conflicting evidence.
Although a significant number of GUS in our cohort have accumulated enough evidence overtime to warrant being reported clinically as pathogenic variants today, the opportunity to do so depends on the initial reporting practice of the laboratory appropriate communication with the clinician, and data reanalysis. Limited information is available on how often, and in what manner, GUSs are disclosed, reanalyzed, and re-reported in practice. With a shortage of genetics professionals, the time required for these efforts may be prohibitive, both in the laboratory as well as clinic. Our data emphasize the need of gene curation, phenotyping, and data sharing in pediatric disorders to increase the diagnostic rate of NGS in individuals with rare diseases. In addition, this study highlights the necessity of close collaboration among ordering physicians, molecular geneticists, and researchers for accurate data interpretation.
ACKNOWLEDGMENTS
We gratefully acknowledge the patients and their families, as well as the referring physicians and thank our colleagues in the Center for Pediatric Genomic Medicine.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
ETHICAL STATEMENT
The project was approved by the ethics committee of Children's Mercy Hospitals.