The ClinGen Epilepsy Gene Curation Expert Panel—Bridging the divide between clinical domain knowledge and formal gene curation criteria
For the ClinGen/ClinVar Special Issue
Abstract
The field of epilepsy genetics is advancing rapidly and epilepsy is emerging as a frequent indication for diagnostic genetic testing. Within the larger ClinGen framework, the ClinGen Epilepsy Gene Curation Expert Panel is tasked with connecting two increasingly separate fields: the domain of traditional clinical epileptology, with its own established language and classification criteria, and the rapidly evolving area of diagnostic genetic testing that adheres to formal criteria for gene and variant curation. We identify critical components unique to the epilepsy gene curation effort, including: (a) precise phenotype definitions within existing disease and phenotype ontologies; (b) consideration of when epilepsy should be curated as a distinct disease entity; (c) strategies for gene selection; and (d) emerging rules for evaluating functional models for seizure disorders. Given that de novo variants play a prominent role in many of the epilepsies, sufficient genetic evidence is often awarded early in the curation process. Therefore, the emphasis of gene curation is frequently shifted toward an iterative precuration process to better capture phenotypic associations. We demonstrate that within the spectrum of neurodevelopmental disorders, gene curation for epilepsy-associated genes is feasible and suggest epilepsy-specific conventions, laying the groundwork for a curation process of all major epilepsy-associated genes.
1 BACKGROUND
Epilepsy is one of the most common brain disorders, affecting up to 3 million people in the United States with an annual cost to the U.S. healthcare system of up to 15 billion USD. Despite the availability of a growing number of antiepileptic medications, up to 30% of persons with epilepsy have treatment-resistant seizures, significantly impacting quality of life and putting patients at risk for various comorbidities and complications including death (Moshe, Perucca, Ryvlin, & Tomson, 2015). Epilepsy can develop in the setting of structural changes to the brain such as injuries or malformations. However, in a significant proportion of patients, no structural alterations can be identified through neuroimaging (Thomas & Berkovic, 2014). Twin studies demonstrate a strong genetic contribution to various epilepsy types, and family studies suggest a strong genetic influence on a population level (Berkovic, Howell, Hay, & Hopper, 1998; Peljto et al., 2014; Vadlamudi et al., 2004). Novel technologies to generate large-scale genetic data have led to various breakthroughs in rare pediatric epilepsies, where precision medicine approaches are already applied (Allen, Berkovic, Cossette, Delanty, & Winawer, 2013 EpiPM Consortium, 2015; EuroEPINOMICS-RES Consortium, Epilepsy Phenome/Genome Project, & Epi4K Consortium, 2017; Reif, Tsai, Helbig, Rosenow, & Klein, 2017).
With the advent of next-generation sequencing technologies, the last decade has seen an explosion of causative genes identified in patients with epilepsy and neurodevelopmental disorders, and currently more than 40 genes are considered bona fide causes of genetic epilepsies, given that pathogenic variants in these genes are identified in patients with epilepsy on a regular basis in a clinical and research setting. Most of these genes are linked to developmental and epileptic encephalopathies, severe epilepsies with an early age of onset, and multiple associated comorbidities. The genetic testing landscape is extremely diverse ranging from targeted testing including single gene assays to exome or genome sequencing. Targeted epilepsy gene panel approaches are equally diverse, with some focusing on bona fide genes causing primary epilepsy and other larger gene panels often including genes related to syndromic disorders or candidate genes related to epilepsy due to their cellular and functional roles.
Given that epilepsy is a dynamic disease occurring over time, it is the hope in the field that genetic findings can be used to guide therapy and improve patient outcomes (EpiPM Consortium, 2015). However, using genetic data for patient treatment requires that genetic findings are systematically vetted for the association with a given disease entity. Although scientific publications on gene discovery have a focus on novelty, there are few mechanisms to track the emerging evidence for genes over time and to systematically assess their validity within a disease context. The Clinical Genome Resource (ClinGen) Gene Curation Expert Panel offers such a mechanism by providing an evidence-based framework to assess the clinical validity of specific gene–disease associations using available genetic and experimental evidence. ClinGen is an NIH-funded initiative dedicated to identifying clinically relevant genes and variants for use in precision medicine and research (Rehm et al., 2015). One of the main tasks of the ClinGen Consortium is the assessment of the validity gene–disease associations, asking the question whether variation in a certain gene has sufficient evidence to be considered causative for a particular phenotype. To this end, the ClinGen Consortium has developed a formal framework to evaluate genetic and experimental evidence supporting or disputing a gene–disease relationship (Strande et al., 2017). This framework will then form the basis to assess variants within these genes based on guidelines, such as the recommendations by the American College of Medical Genetics and Genomics (Richards et al., 2015). Deposition of variants in curated genes in public archives such as ClinVar (Landrum et al., 2016) will then allow variant information to be used in diagnostic and research settings. However, prior to considering evidence for a particular variant and considering potential actionability of genetic testing, sufficient evidence for the involvement of a gene within the context of a particular disease needs to be established.
The ClinGen Epilepsy Gene Curation Expert Panel is tasked with assessing the validity of gene–disease associations related to human epilepsy within the formal, evidence-based gene curation framework of the wider ClinGen Consortium, with the ultimate goal that these findings will inform future decisions about gene selection for diagnostic tests and future studies into precision medicine approaches. In parallel, the epilepsy field has a rich tradition in studying genetic causes of human epilepsy that is traditionally focused on phenotyping. The current manuscript describes the pilot activities during the first year of the ClinGen Epilepsy Gene Curation Expert Panel, with an emphasis on harmonization between traditional clinical epilepsy concepts and the ClinGen framework to lay the groundwork for a larger gene curation effort in the future.
2 METHODS AND RESULTS
2.1 Composition of the ClinGen Epilepsy Gene Curation Expert Panel
The ClinGen Epilepsy Gene Curation Expert Panel has been active since June 2017 and consists of a mixture of clinical epileptologists, medical geneticists, genetic counselors, clinical molecular geneticists, basic scientists, and biocurators. The composition of the working group is international, with members from the United States, Europe, and Canada. The ClinGen Epilepsy Gene Curation Expert Panel is embedded within the ClinGen Neurodevelopmental Disorders Clinical Domain Working Group (CDWG) that also includes the Intellectual Disability/Autism Gene Curation Expert Panel and the Rett/Angelman-like disorders Variant Curation Expert Panel. Two epilepsy gene specific variant curation expert panels are planned as future components of the Neurodevelopmental Disorders CDWG including the KCNQ2 Expert Panel and the NMDA receptor Variant Curation Expert Panel (GRIN1, GRIN2A, GRIN2B, GRIN2D). The ClinGen Epilepsy Gene Curation Expert Panel is also affiliated with the International League against Epilepsy (ILAE) Genetics Commissions through prior membership in the Genetics Commission during the 2014—2017 term for Ingo Helbig and Heather Mefford and current affiliation for the 2017–2020 term through the “Epilepsiome” Task Force, linking the ClinGen gene curation activities with the genetic literacy series of the ILAE Genetics Commission (Helbig, Heinzen, Mefford, & ILAE Genetics Commission, 2016; Tan, Lowenstein, & ILAE Genetics Commission, 2015) and peer-to-peer communication through a dedicated blog (“Beyond the Ion Channel”; epilepsygenetics.net).
2.2 Strategies of gene selection
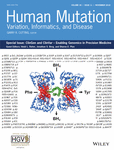
Within the expert panels of the Neurodevelopmental Disorders CDWG, various strategies were used to select a set of genes for initial curation (Figure 1). Given the frequent use of dedicated gene panels in a clinical setting, the epilepsy expert panel decided to focus on a limited number of genes as the first goal of gene curation, curating the “average gene panel” used in clinical practice. In order to identify commonly tested genes, we compiled a list of 2,702 genes from 236 commercial gene panels through a query of the Genetic Testing Registry for tests with the keywords “seizure OR epilepsy” (Rubinstein et al., 2013). In order to select genes with a primary epilepsy phenotype as opposed to genes with epilepsy as a contributing feature, we compiled a condensed gene list of 123 genes, combining evidence from literature (Heyne et al., 2018; Lindy et al., 2018), and expert opinion. Of this gene list, 29 genes were selected for the pilot phase and for the precuration process, a review of published phenotypes prior to initiating the formal curation process (discussed below). Genes with primary syndromic or nonepilepsy neurodevelopmental phenotypes including autism and intellectual disability were excluded, such as ZEB2 for Mowat–Wilson syndrome. In addition, genes primarily associated with a Rett-like phenotype such as MECP2 or CDKL5 were removed from the gene list, given the existence of a dedicated working group for these conditions within ClinGen. For some genes, an iterative review after a primary curation has revealed that further precuration is required to determine the clinical validity of gene for a specific epilepsy phenotype (e.g., GRIN1). The list of precurated genes will then undergo a formal gene curation process after selecting and possibly refining an adequate epilepsy phenotype within the Monarch Disease Ontology (MONDO; Figure 2). The pilot phase of the ClinGen Epilepsy Expert Panel highlighted issues related to traditional clinical epilepsy classification for which we developed an iterative process to systematically curate epilepsy-associated genes, which is currently in process.
2.3 Piloting epilepsy gene curation
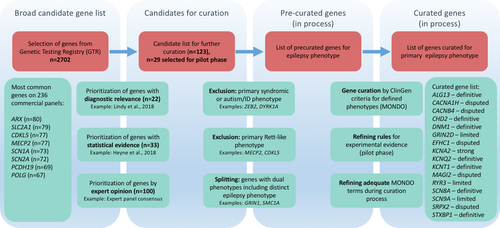
The ClinGen gene curation framework uses a dedicated gene curation interface (GCI) and (MONDO for the specification of the disease entity of interest. During the first year, an iterative process was adopted to initiate gene curation on a few selected genes to evaluate the ClinGen gene curation framework and its dedicated tools as they apply specifically to the epilepsies. The pilot evaluation phase led to a process of curation and precuration and an interactive process of defining the most appropriate grouping of phenotypes for downstream gene curation. Using the previously described evidence-based framework from the ClinGen Gene Curation Working Group (Strande et al., 2017), we evaluated the clinical validity for the proposed gene–disease relationships for 16 genes (Table 1). The ClinGen clinical validity framework uses two main classes of available evidence, both genetic and experimental, to derive a semiquantitative measurement of the strength of the evidence for gene–disease associations. The classifications of the strength of the gene–disease relationship include: “Definitive,” “Strong,” “Moderate,” “Limited,” “No Reported Evidence,” and “Disputed.” In order to achieve a classification of “Definitive,” a gene–disease association must achieve at least 12 points and demonstrate replication over time, which is defined as at least two publications reporting pathogenic variants in the gene and at least 3 years since the initial report. Our curation of the initial 16 genes resulted in a classification as “Definitive” gene–disease association in seven of 16 genes, “Strong” in one of 16 genes, “Limited” in three of 16 genes, and “Disputed” in five of 16 genes (Table 1).
| Gene | Disease entity | Date of curation | Genetic evidence (points) | Experimental evidence (points) | Total points | Replication over time | Classification |
|---|---|---|---|---|---|---|---|
| ALG13 | Undetermined Early-Onset Epileptic Encephalopathy (MONDO:0018614) | 3/14/18 | 12 | 0 | 12 | Yes | Definitive |
| CHD2 |
|
7/18/17 | 12 | 3 | 15 | Yes | Definitive |
| DNM1 | Infantile Epilepsy Syndrome (MONDO:0020071) | 5/31/18 | 12 | 4.5 | 16.5 | Yes | Definitive |
| KCNQ2 | Early Infantile Epileptic Encephalopathy (MONDO:0016021) | 9/5/17 | 12 | 5.5 | 17.5 | Yes | Definitive |
| KCNT1 |
|
7/26/17 | 12 | 2 | 14 | Yes | Definitive |
| SCN8A | Infantile Epilepsy Syndromea (MONDO:0020071) | 1/6/17 | 12 | 6 | 18 | Yes | Definitive |
| STXBP1 | Early Infantile Epileptic Encephalopathya (MONDO:0016021) | 6/15/17 | 12 | 6 | 18 | Yes | Definitive |
| KCNA2 | Infantile Epilepsy Syndrome (MONDO:0020071) | 10/5/17 | 12 | 4 | 16 | No | Strong |
| GRIN2D | Infantile Epilepsy Syndrome (MONDO:0020071) | 7/3/18 | 3 | 1.5 | 4.5 | No | Limited |
| RYR3 | Undetermined Early Onset Epileptic Encephalopathy (MONDO:0018614) | 6/28/18 | 1 | 0 | 1 | No | Limited |
| SCN9A | Epilepsy (MONDO:0005027) | 6/15/18 | 3.8 | 0.5 | 4.3 | No | Limited |
| CACNA1H | Generalized Epilepsy (MONDO:0005579) | 7/31/18 | 0 | 4.5 | 4.5 | No | Disputed |
| CACNB4 | Generalized Epilepsy (MONDO:0005579) | 6/22/18 | 0 | 2 | 2 | No | Disputed |
| EFHC1 | Juvenile Myoclonic Epilepsy (MONDO:0009696) | 7/27/18 | 0 | 1 | 1 | No | Disputed |
| MAGI2 | Infantile Epilepsy Syndrome (MONDO:0020071) | 6/26/18 | 0 | 0.5 | 0.5 | No | Disputed |
| SRPX2 | Rolandic Epilepsy-Speech Dyspraxia Syndrome (MONDO:0015587) | 7/19/18 | 0 | 0 | 0 | No | Disputed |
- a Provisionally curated for this term with the understanding that it does not fully encompass the range of phenotypes associated with the disease; in the future, “Complex Neurodevelopmental Disorder” will be used as disease entity.
- b During the precuration phase, it was decided to consider Epilepsy of Infancy with Migrating Focal Seizures and Autosomal Dominant Nocturnal Frontal Lobe Epilepsy as one disease entity, since families have been reported with individuals with both clinical presentations, and the same pathogenic KCNT1 variant has been associated with both clinical presentations. MONDO:0020072 has been used as a temporary placeholder.
2.4 Genetic evidence
Within the ClinGen gene curation framework, genetic evidence is derived from publicly available data describing variants in the gene of interest identified in patients with the disease entity of interest. Genetic evidence is divided into two categories: (a) case-level data, in which studies report individuals or families with genetic variants; and (b) case-control data, which is derived using statistical analyses in case-control studies. A maximum score of 12 points can be achieved through genetic evidence, with points awarded for variant segregation and type for case-level data, and methodology, statistical power, bias and confounding factors, and statistical significance for case-control data. Inheritance pattern within reported cases is a strong consideration when assessing available case-level genetic evidence, and in this regard the underlying genetic architecture of early-life epilepsies, with a strong contribution of de novo variants, lends itself to achieving a high number of points for case-level genetic evidence. Eight of 16 genes in our pilot curation phase achieved maximum genetic evidence of 12 points within the existing ClinGen gene curation framework, which suggests that sufficient genetic evidence according to ClinGen criteria is easily achieved both for well-established genetic causes of epilepsy including SCN8A and KCNQ2 and more recently implicated genes such as KCNA2 and ALG13 (Supporting Information Table 1). However, we also identified eight genes with limited or disputed evidence, suggesting that some of the genes traditionally considered genetic etiologies for epilepsy have limited or even contradictory evidence. For example, genes such as EFHC1 or CACNA1H are disputed by formal ClinGen criteria, despite the fact that these genes are part of currently available diagnostic gene panels (EFHC1 n = 33 gene panels; CACNA1H n = 13 gene panels). Most genetic epilepsies have been described within the last 5 years in next-generation sequencing studies, allowing for comparison of variant frequencies in patients with population databases such as ExAC and gnomAD (Lek et al., 2016), which can be used as control populations for severe early-onset epilepsies. Therefore, minor allele frequencies in control populations, segregation, and absence of other explanatory genetic etiologies are frequently available and not a limiting factor in assessing gene validity within the epilepsies. Within the context of the traditional clinical concept of genetic epilepsies, the existing ClinGen clinical validity criteria were found to be adequate and to be reflective of the general consensus in the epilepsy field.
During our initial pilot curation phase, the expert panel noted that within the epilepsies, the development of gene-specific assessment criteria may be helpful, both for gene curation and variant interpretation. For example, a majority of known genetic epilepsies are considered “channelopathies,” resulting from pathogenic variants in neuronal ion channel encoding genes, often with a gain-of-function effect (Oyrer et al., 2018). Consequently, loss-of-function variants, which are typically considered damaging or pathogenic in most genes, may actually be tolerated in many neuronal ion channels or result in milder phenotypes, as is the case in KCNQ2 (Miceli et al., 1993). Domain knowledge, both of expected clinical phenotypes for genetic epilepsy syndromes as well as expected variants and functional consequences, is essential in appropriately interpreting the significance of epilepsy-associated variants. A future goal of the ClinGen Epilepsy Gene Curation Expert Panel is to develop gene-specific assessment criteria in the context of the epilepsies, taking these and other considerations into account, which will aid in curation both of gene-disease associations as well as variant interpretation.
The expert panel has not yet considered gene–disease relationships where gene validity was primarily asserted through association studies. Evaluation of case-control data will be particularly relevant for milder, complex genetic epilepsies including the genetic generalized epilepsies and nonlesional focal epilepsies, where monogenic factors play a role in a minority of patients. Future curation efforts taking into account genetic evidence derived from case-control studies will allow the expert panel to assess its applicability to epilepsies with a complex underlying genetic architecture.
2.5 Experimental evidence
Gene-level experimental evidence is derived within the ClinGen framework by assessing the following types of available evidence: biochemical function, experimental protein interactions, expression, functional alteration in patient and nonpatient derived cells, phenotypic rescue, and animal model systems (Strande et al., 2017). A maximum of six points can be achieved from experimental evidence, taking various factors into account including, but not limited to, the relevance and robustness of the experimental assay. Experimental evidence in the eight genes with strong or definite evidence assessed during the pilot curation phase ranged from absent to the full amount of six possible points (Table 1). Only one gene–disease association was awarded no points for experimental evidence, due to lack of available evidence. The remaining seven genes were awarded some experimental evidence points, ranging from two to six points. The types of experimental evidence used to assess the gene–disease associations were highly variable, including biochemical function, expression, functional alteration (largely in nonpatient cells), and animal models (Supporting Information Table 1). Although the neuroscience field has a strong tradition of functional studies in neuronal ion channels (Oyrer et al., 2018), the pilot curation phase did not reflect a bias toward experimental evidence for ion channel genes; experimental evidence points were also awarded for non-ion channel encoding genes including DNM1, CHD2, and STXBP1, although notably not ALG13. However, experimental evidence was not needed to obtain sufficient points for the classification of genes as definitive or strong for any of the eight genes with strong or definite evidence curated within our pilot phase. This indicates that the evidence for gene validity in the epilepsy field is primarily driven by genetic findings due to the large number of published studies and a high proportion of genetic epilepsies due to de novo variants and thus generally do not require experimental evidence as support.
The ClinGen gene curation framework is used as a guide, and in certain scenarios it is appropriate for the expert panel to adjust scoring or final classifications based on professional judgment. Given this consideration and the complexity of neurodevelopmental phenotypes, the expert panel decided to award reduced points for experimental evidence in some scenarios. For example, for KCNA2, the main mouse model has an ataxia phenotype but not a seizure phenotype (Xie et al., 2010). Although ataxia is increasingly recognized as a common feature in patients with KCNA2-related neurodevelopmental disorders, the question arises how related phenotypic features should be scored within the concept of primarily assessing functional evidence toward the epilepsy phenotype. Within the working group, it was agreed that the presence of incomplete neurological phenotypes in model systems, which may include movement disorders, would be scored at 0.5 compared to the default of 2 points for animal models that exhibit spontaneous seizures.
2.6 Use of disease and phenotype ontologies
Within the ClinGen framework, curation for a gene–disease association requires the selection of a disease entity for which a given gene is curated. Disease entities within the ClinGen framework are coded within the MONDO (https://www.ebi.ac.uk/ols/ontologies/mondo), an aggregate ontology of human disease phenotypes using a hierarchical concept of parent and child terms (Figure 2). Although MONDO includes epilepsy syndromes, the human phenotype ontology (HPO) is an ontology of symptoms, which, in the context of epilepsies, would include seizure types and defined comorbidities such as intellectual disability or movement disorders. Epilepsy is a field with a rich tradition focused on electroclinical phenotyping, and the concept of using disease and phenotype ontology is relatively new. Only some research initiatives such as the EuroEPINOMICS-RES Consortium have consistently used HPO terms and have been involved in the generation of these ontologies (Kohler et al., 2014, 2017).
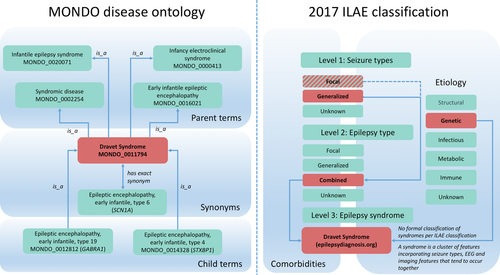
Disease and phenotype ontologies have often been generated through a computational data aggregation process that may result in inconsistencies with existing clinical classifications at the level of individual disease phenotypes. We identified a lack of correspondence of known disease entities in MONDO with the current and previous ILAE classification of the epilepsies (Scheffer et al., 2017), indicating the need to align the MONDO with classifications that are used clinically and in epilepsy genetics research, such as the 2017 ILAE (Figure 2). We identified concepts within the ILAE classification that cannot be easily translated into a disease ontology primarily defined by phenotypic features such as MONDO. Although many epilepsy syndromes can be mapped onto the MONDO, the classification of epilepsy by etiology, for instance, cannot be easily translated to the MONDO. Figure 2 demonstrates the differences in classification for Dravet syndrome (MONDO_0011794). Within the 2017 ILAE classification, seizure types, epilepsy types, and epilepsy syndromes are classified on different levels, whereas the MONDO provides various parent terms for Dravet syndrome, reflecting the use of this epilepsy syndrome in various contexts. Within the ILAE classification, epilepsy syndromes are referred to as clusters of clinical, electroencephalography (EEG), or imaging features, but once a diagnosis of a specific syndrome is made it is not defined to which broader parent term the syndrome belongs to. However, such a hierarchical structure is the basis of the MONDO.
2.7 Iterative curation of epilepsy-related genes
2.7.1 Precuration
In complex neurodevelopmental disorders, the review of associated phenotypes led to an iterative process of curation including a precuration step. The ClinGen Lumping and Splitting Working Group has developed precuration guidelines that have been used by the epilepsy expert panel in precuration efforts during our pilot phase (https://www.clinicalgenome.org/working-groups/lumping-and-splitting/). The precuration process includes a review of published phenotypes prior to launching the gene curation process, leading to a possible assertion of distinct phenotypes versus disease spectrums, which is then substantiated or refuted during the precuration review of the evidence for or against distinct phenotypes. For a range of disease genes, a spectrum of disease entities has been observed, sometimes even with distinct disease entities associated with identical variants.
During the curation of the 16 epilepsy-related genes and selection of 27 additional genes for precuration in the pilot phase, we observed that traditional clinical distinctions between known disease entities may not necessarily apply when using the ClinGen framework for genetic etiologies associated with epilepsy. A “variant first” approach to lumping and splitting of epilepsy phenotypes is conceptually different from the “phenotype first” approach in a clinical setting, and it may not be sensitive to known distinctions between clinical entities if the variants overlap. For example, in the case of SCN1A-related disorders, there is a traditional clinical distinction between Dravet syndrome, a distinct developmental and epileptic encephalopathy that presents in the first year of life, and other forms of generalized epilepsy as a milder phenotype (Steel, Symonds, Zuberi, & Brunklaus, 2017; Zhang et al., 2017). However, as there are at least some families reported with overlapping phenotypes associated with the same variant (Goldberg-Stern et al., 2014; Hoffman-Zacharska et al., 2015), SCN1A-related disorders would be primarily considered a spectrum and would be curated for a broad rather than narrow phenotype. Similar observations were made for genes such as SCN2A and SCN8A. Alternatively, some gene-disease associations emerged as distinct phenotypes that were less apparent at the outset. For example, in ALG13, strong evidence emerged for an epilepsy phenotype in females with a recurrent de novo variant. However, only limited evidence arose for the congenital disorders of glycosylation phenotype that was first described and that is consistent with the presumed function of this gene. Due to the accrual of genetic evidence through multiple reports, the female epileptic encephalopathy phenotype that was initially considered a subphenotype has “overtaken” the initial ALG13 phenotype with respect to gene validity. Finally, some genes behaved as expected in the precuration process. For example, for KCNQ2, precuration successfully identified both the mild phenotype due to haploinsufficiency and the more severe phenotype primarily associated with missense variants with predicted dominant-negative effect, mirroring the separation between known clinical entities, self-limited neonatal seizures (also known as benign familial neonatal epilepsy), and KCNQ2 encephalopathy, within the ClinGen precuration framework.
3 DISCUSSION
In the current manuscript, we describe the pilot phase of the epilepsy gene curation activities within the ClinGen Epilepsy Gene Curation Expert Panel. We demonstrate that the established gene curation process can be applied to genetic etiologies linked to human epilepsy with special considerations. We observe that genetic evidence for a selection of epilepsy genes in the pilot phase can be readily provided through the published literature. Both the de novo architecture of neurodevelopmental disorders as well as the high frequency of follow-up publications focusing on phenotype delineation contribute to this effect. However, other genes frequently tested on diagnostic gene panels have contradictory evidence, and the gene–disease relationships must be considered disputed by the formal criteria of the ClinGen Consortium (CACNA1H, CACNB4, EFHC1, MAGI2, SRPX2). Three genes curated within the pilot phase have limited evidence (GRIN2D, RYR3, SCN9A), indicating that more evidence is needed to support a strong or definite gene–disease association within the context of epilepsy. We identify the appropriate selection of the disease phenotype as one of the major challenges in the curation effort, an activity that is usually referred to as precuration.
In contrast to many other disease entities, the epilepsies are phenotypes that are not easily classified within the existing ontologies and format, requiring the ClinGen Epilepsy Gene Curation Expert Panel to put a strong focus on the precuration effort. Given that both the MONDO and HPO classifications are increasingly used in a diagnostic context, we highlight the importance of iteratively improving existing ontologies to align these classifications with the classifications used in a clinical setting. In other words, classifications used for diagnosing genetic epilepsies should harmonize with the schema used by the epileptologists who treat these patients.
Within the formal ClinGen framework, assessment of gene validity precedes the interpretation on the variant level according to variant classification guidelines such as the American College of Medical Genetics and Genomics (ACMG) recommendations. However, particularly for the well-studied ion channel genes, identified variants have been demonstrated to have variable, if not, opposite functional effects. For example, disease-causing gain-of-function and loss-of-function variants are observed in genes such as SCN2A or SCN8A. This observation raises the issue whether the variant-level interpretation can be separated from the gene-level interpretation. The ClinGen Consortium has addressed this question by providing recommendations on when phenotypes linked to a particular gene should be lumped into a single phenotypic spectrum or split into separate phenotypes (https://www.clinicalgenome.org/working-groups/lumping-and-splitting/). The identification of appropriate phenotypes is part of the precuration effort, further emphasizing the need for a detailed precuration phase for epilepsy-related genes. We have assessed this question for SCN8A where both gain-of-function and loss-of-function variants have been described in the literature. We concluded that the SCN8A-related disorders demonstrate a broad spectrum independent of the functional effect of the variant, including variable presentations for known recurrent variants. With increasing knowledge about different phenotypes, outcomes, and therapeutic responses, some of the curated genes may have sufficient evidence to be split into distinct phenotypes in the future.
Harmonizing traditional epilepsy phenotypes with the phenotypic categories provided by the MONDO provided a particular challenge and for some phenotypes, the existing disease ontologies were insufficient. For example, the diagnostic term Early Infantile Epileptic Encephalopathy is increasingly used both in clinical and diagnostic settings, but the term as defined by the MONDO disease ontology does not match the accepted clinical definition for this term. In order to overcome the present mismatches between existing clinical classifications and MONDO, the members of the ClinGen Epilepsy Gene Curation Expert Panel have agreed on using specific terms such as “Early Infantile Epileptic Encephalopathy” (MONDO ID:0016021) as placeholder terms for agreed-upon clinical concepts such as Developmental and Epileptic Encephalopathy despite some inconsistency of parent and child terms within the current ontology while ongoing collaboration with the MONDO Consortium continues in order to better define epilepsy-related syndromes that resemble existing clinical classifications such as the ILAE classification within the MONDO etiology. We recognize that the term “Epileptic Encephalopathy” as it is used for our gene curation purposes is an imperfect placeholder and does not reflect the full clinical spectrum of the genetic epilepsies that have been curated, nor does it necessarily accurately reflect the clinical concept of an epileptic encephalopathy (Howell, Harvey, & Archer, 2016). However, given that the MONDO disease ontology is interlinked with corresponding HPO terms that are used in many diagnostic laboratories to define the phenotypic overlap of specific genetic variants, aligning clinical classifications with ontologies used in laboratory diagnostics is an important prerequisite for meaningful gene and variant interpretation in a diagnostic setting.
The corollary of this process is that the task of the Epilepsy Gene Curation Expert Panel is expanding, shifting from a traditional gene curation platform to an initiative to systematize the representation of epilepsy-related terminology in disease and phenotype ontologies that will provide the basis for bioinformatic assessments of phenotypic overlaps. The iterative process of refining ontological entities prior to gene curation is unique to the ClinGen Epilepsy Gene Curation Expert Panel and reflects the traditional focus on phenotyping within the epilepsy field.
Going forward, the ClinGen Epilepsy Gene Curation Expert Panel will curate all major genes related to human epilepsies in a systematic fashion to suggest gene-specific variant curation criteria that will help reduce the high burden of variants of uncertain significance. This will aid the ultimate goal of providing a framework for accurately assessing variant pathogenicity for future precision medicine interventions.
ACKNOWLEDGMENTS
The ClinGen Epilepsy Gene Curation Expert Panel would like to acknowledge the assistance and support of Kristy Lee, who served as previous coordinator for the expert panel. The ClinGen consortium is funded by the National Human Genome Research Institute of the National Institutes of Health through the following grants and contracts: U41HG006834 (Rehm), U41HG009650 (Berg), 01HG007437 (Berg).
CONFLICTS OF INTEREST
KMK reports personal fees from UCB, Eisai, Novartis, and GW pharmaceuticals outside the submitted work. KL is an employee of Quest (Athena) Diagnostics. EB is an employee of GeneDx. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.