Journal list menu
Export Citations
Download PDFs
Cover Picture
Cover Image:
- Page: C1
- First Published: 16 August 2024
The Sapindaceae family, also known as the soapberry family, comprises over 140 genera and approximately 1,900 species, including economically important and popular fruit trees like lychee, longan, rambutan, and ackee; timber trees as the maple and buckeye; and other species that are prized for their abundant secondary metabolites, such as saponins from soapberry and seed oil from yellowhorn. The cover features the letters “SAP”, representing the Sapindaceae genome database SapBase, filled in with images of key species within the Sapindaceae. SapBase is an integrative genomic resource and analysis platform for the Sapindaceae family established by Li et al. (pages 1561–1570). SapBase provides a critical foundation for research on the diverse species within the Sapindaceae.
Issue information page
Commentary
Rice seed germination priming by salicylic acid and the emerging role of phytohormones in anaerobic germination
- Pages: 1537-1539
- First Published: 24 June 2024
This Commentary examines a recent study that identified peroxisomal cinnamate: CoA ligases as key enzymes for salicylic acid biosynthesis, which promotes submerged germination by releasing the inhibition of rice germination by indole-acetic acid. This study thus provides important information for developing rice varieties suitable for direct seeding.
Brief Communications
Nullification of GFTs fortifies bioactive folates in foxtail millet
- Pages: 1540-1543
- First Published: 02 May 2024
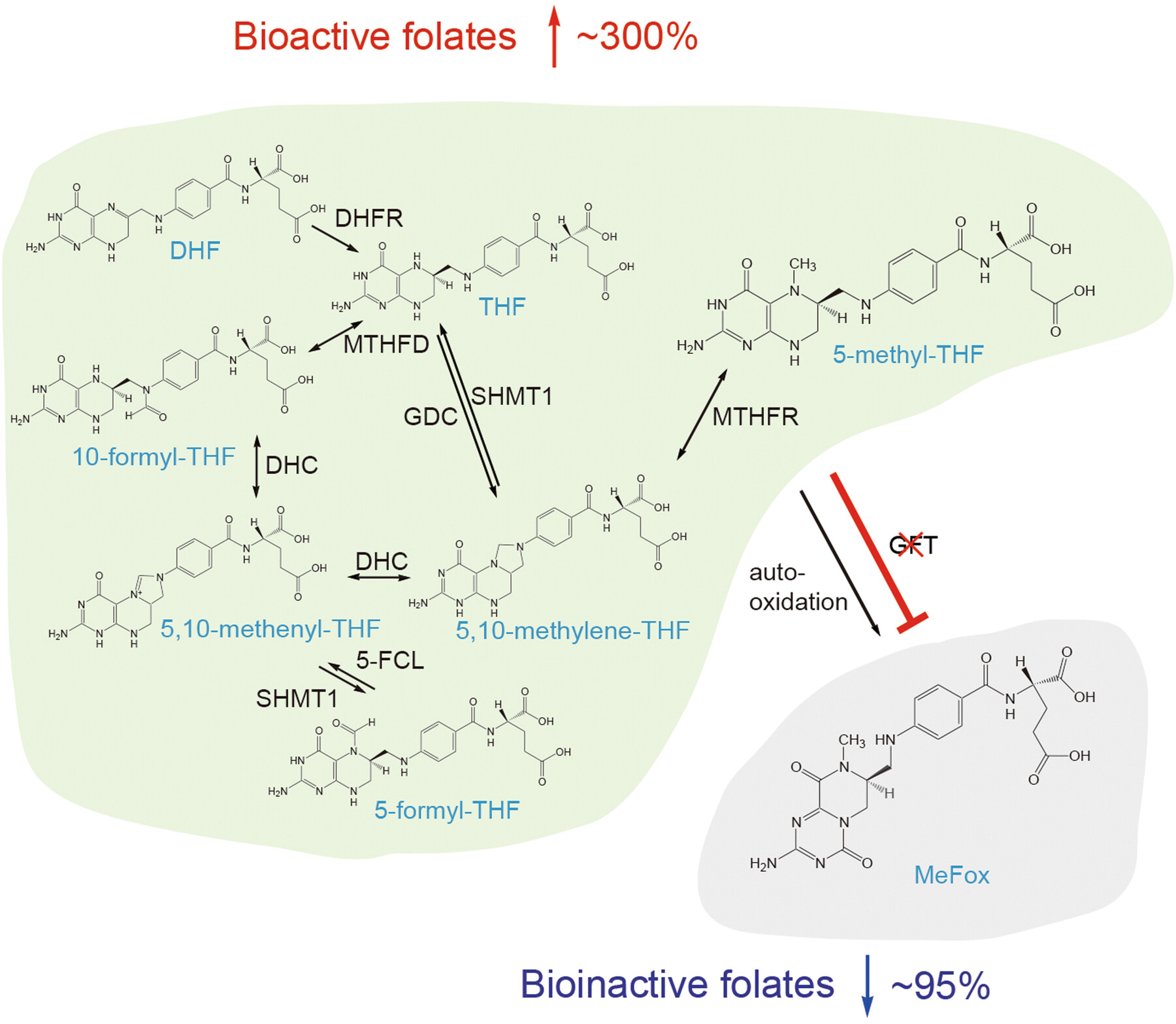
In foxtail millet (Setaria italica), knockout of the glutamate formiminotransferases SiGFT1 and 2 increased the accumulation of bioactive folates to approximately four times the level of wild-type plants and decreased levels of the bioinactive oxidation product MeFox by 95%, thus providing a promising route for folate biofortification in cereal crops.
Application of CRISPR/Cas12i.3 for targeted mutagenesis in broomcorn millet (Panicum miliaceum L.)
- Pages: 1544-1547
- First Published: 02 May 2024
Oomycete Nudix effectors display WY-Nudix conformation and mRNA decapping activity
- Pages: 1548-1552
- First Published: 18 June 2024
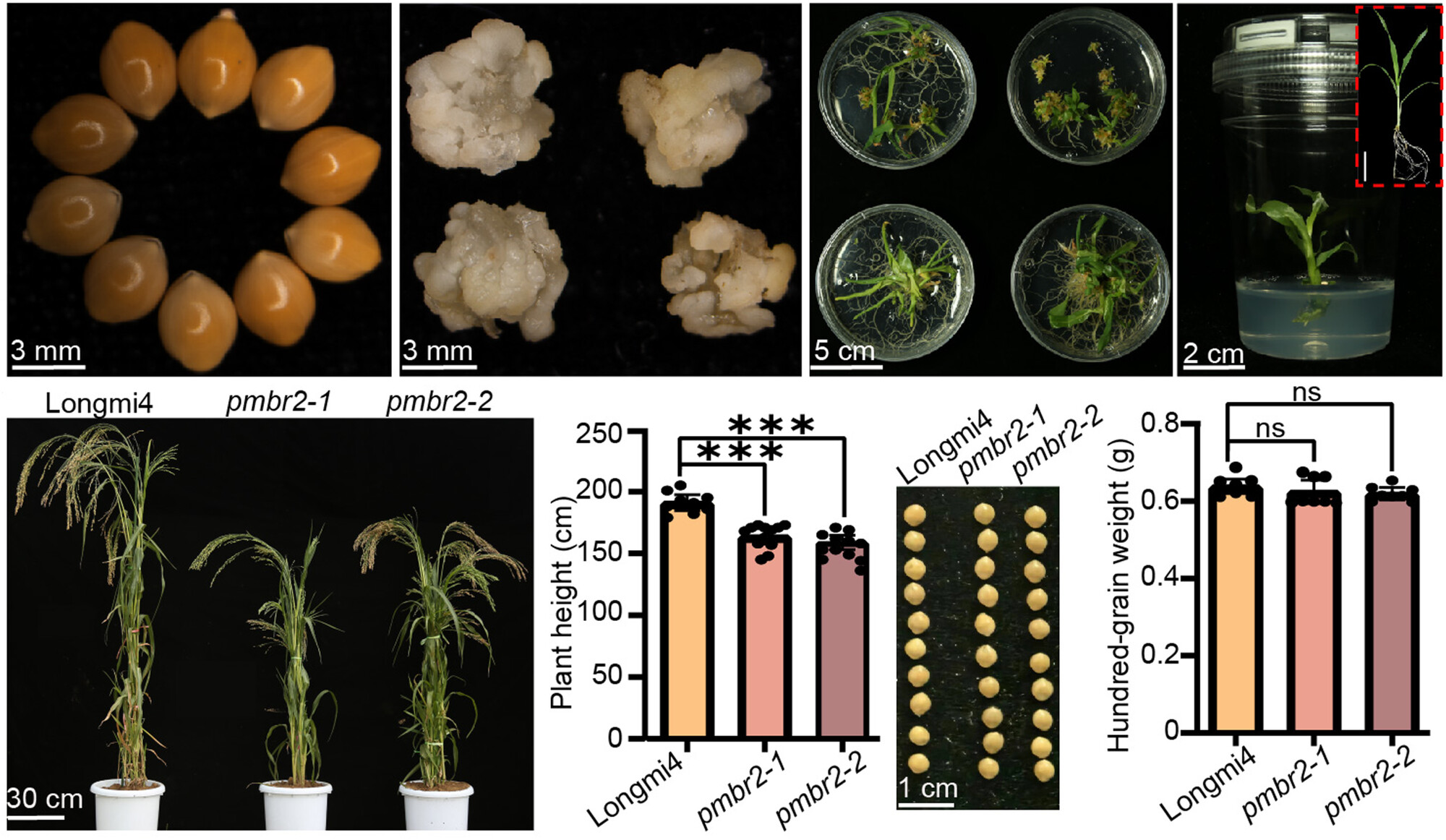
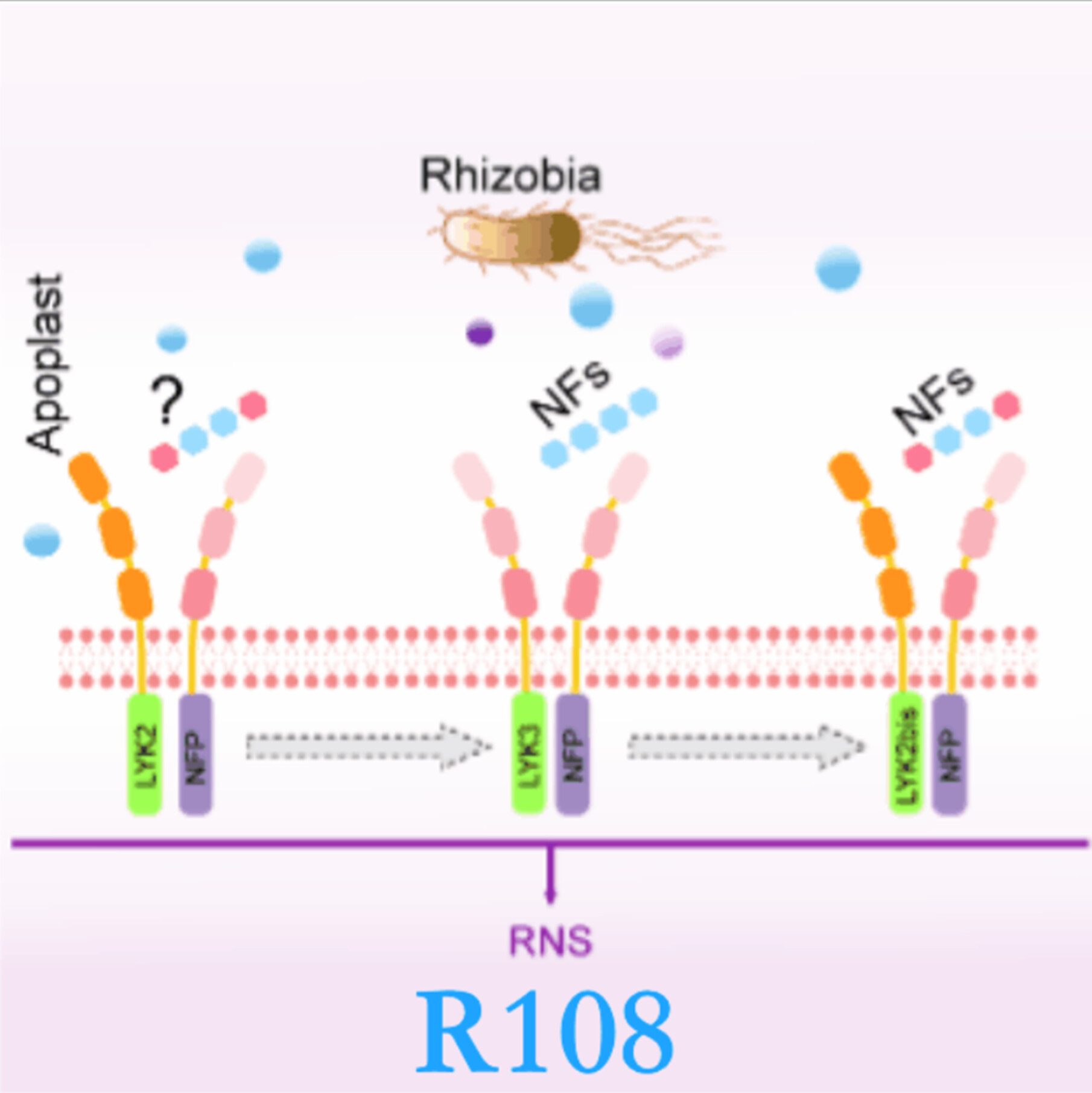
NODULATION TRIO in Medicago truncatula: Unveiling the redundant roles of MtLYK2, MtLYK3, and MtLYK2bis
- Pages: 1553-1556
- First Published: 18 June 2024
Developing guanine base editors for G-to-T editing in rice
- Pages: 1557-1560
- First Published: 27 June 2024
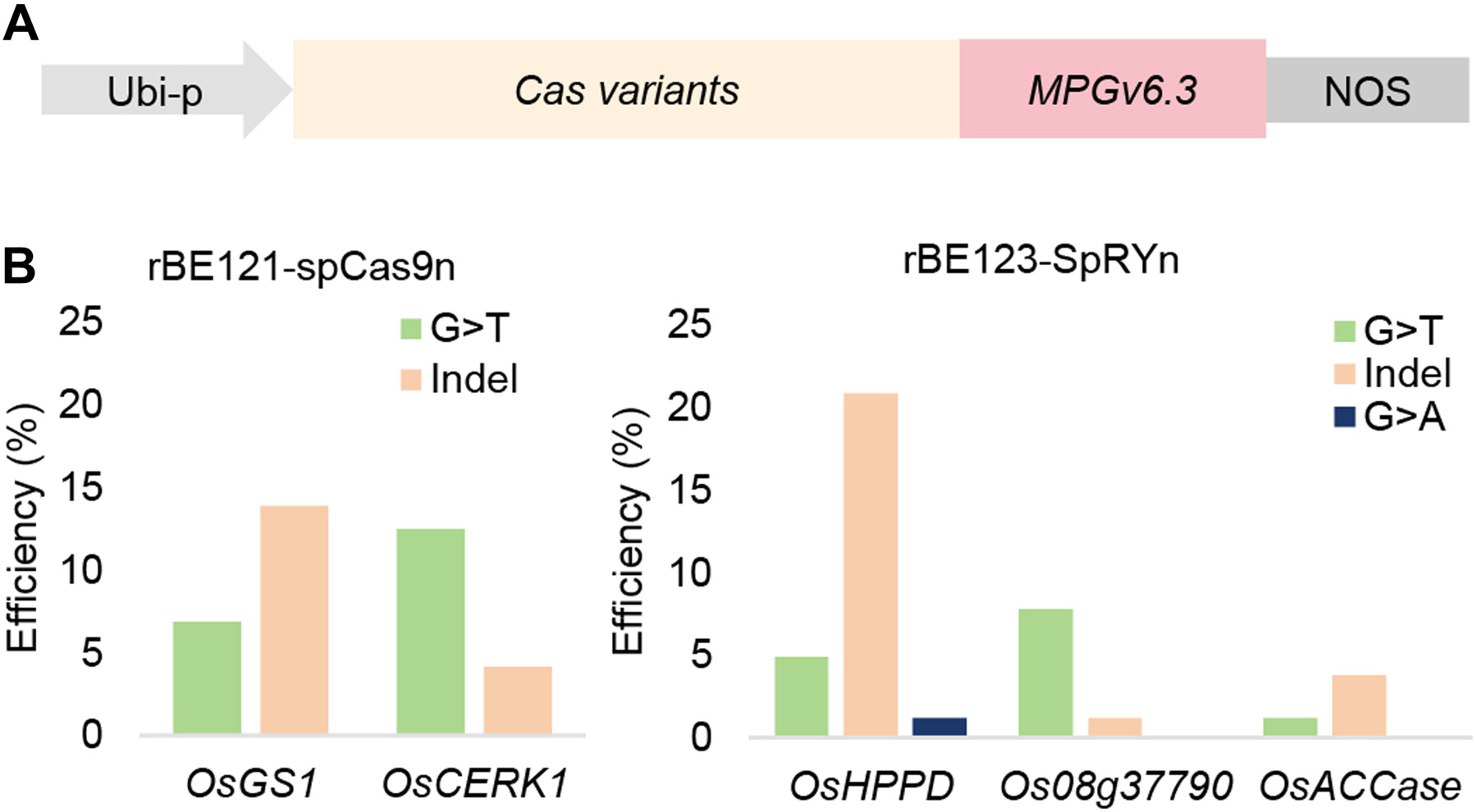
Two guanine base editors created using an engineered N-methylpurine DNA glycosylase with CRISPR systems achieved targeted G-to-T editing with 4.94–12.50% efficiency in rice (Oryza sativa). The combined use of the DNA glycosylase and deaminases enabled co-editing of target guanines with adenines or cytosines.
New Resources
SapBase: A central portal for functional and comparative genomics of Sapindaceae species
- Pages: 1561-1570
- First Published: 28 May 2024
SapBase, a comprehensive, one-stop resource and analysis platform dedicated to the Sapindaceae family, aggregates publicly available genomes and omics data from seven Sapindaceae species, thus providing user-friendly tools for gene information mining, co-expression analysis, and comparative genomic analysis across species.
Abiotic Stress Responses
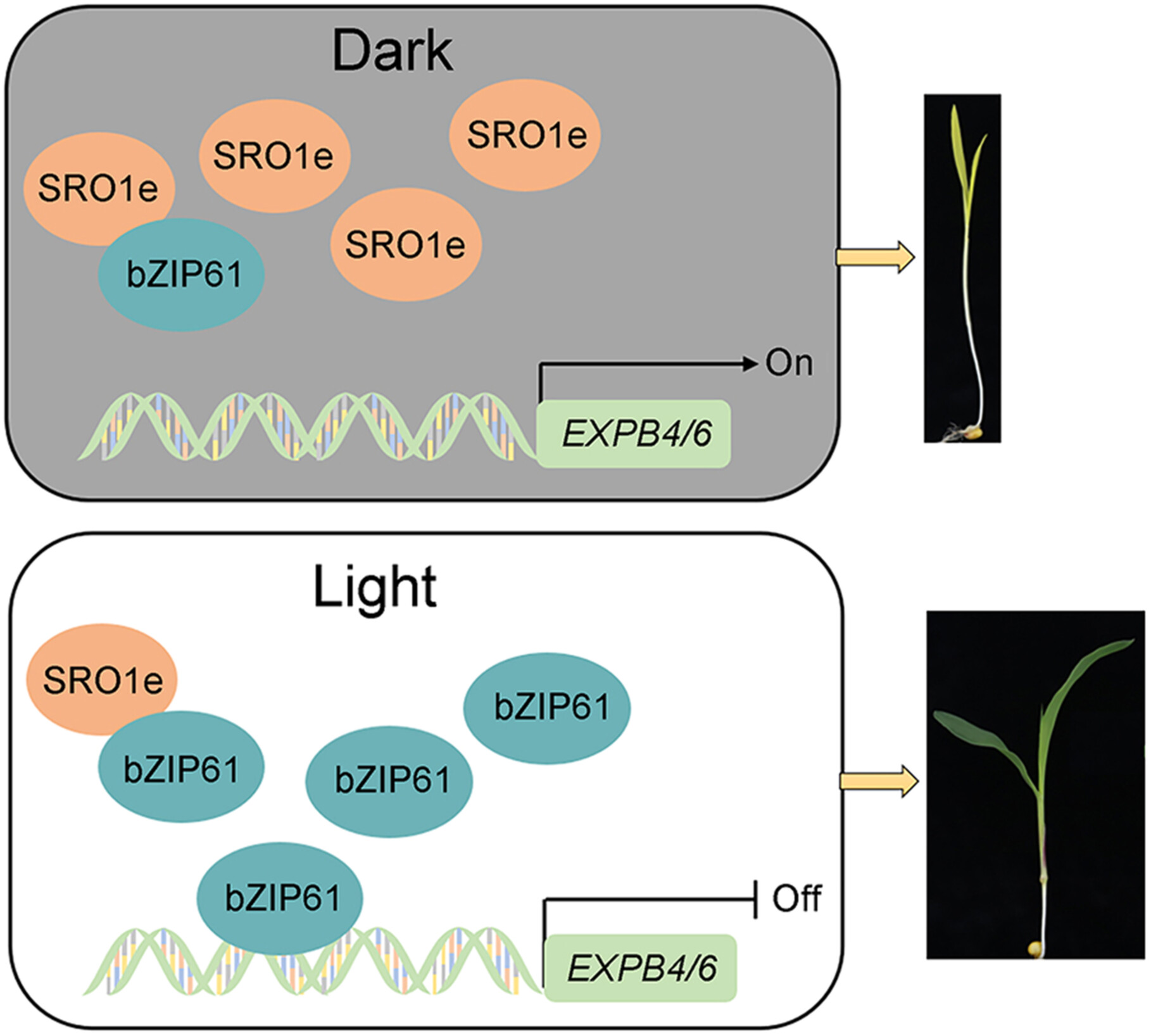
Maize ZmSRO1e promotes mesocotyl elongation and deep sowing tolerance by inhibiting the activity of ZmbZIP61
- Pages: 1571-1586
- First Published: 14 June 2024
Overexpression of tonoplast Ca2+-ATPase in guard cells synergistically enhances stomatal opening and drought tolerance
- Pages: 1587-1602
- First Published: 24 June 2024
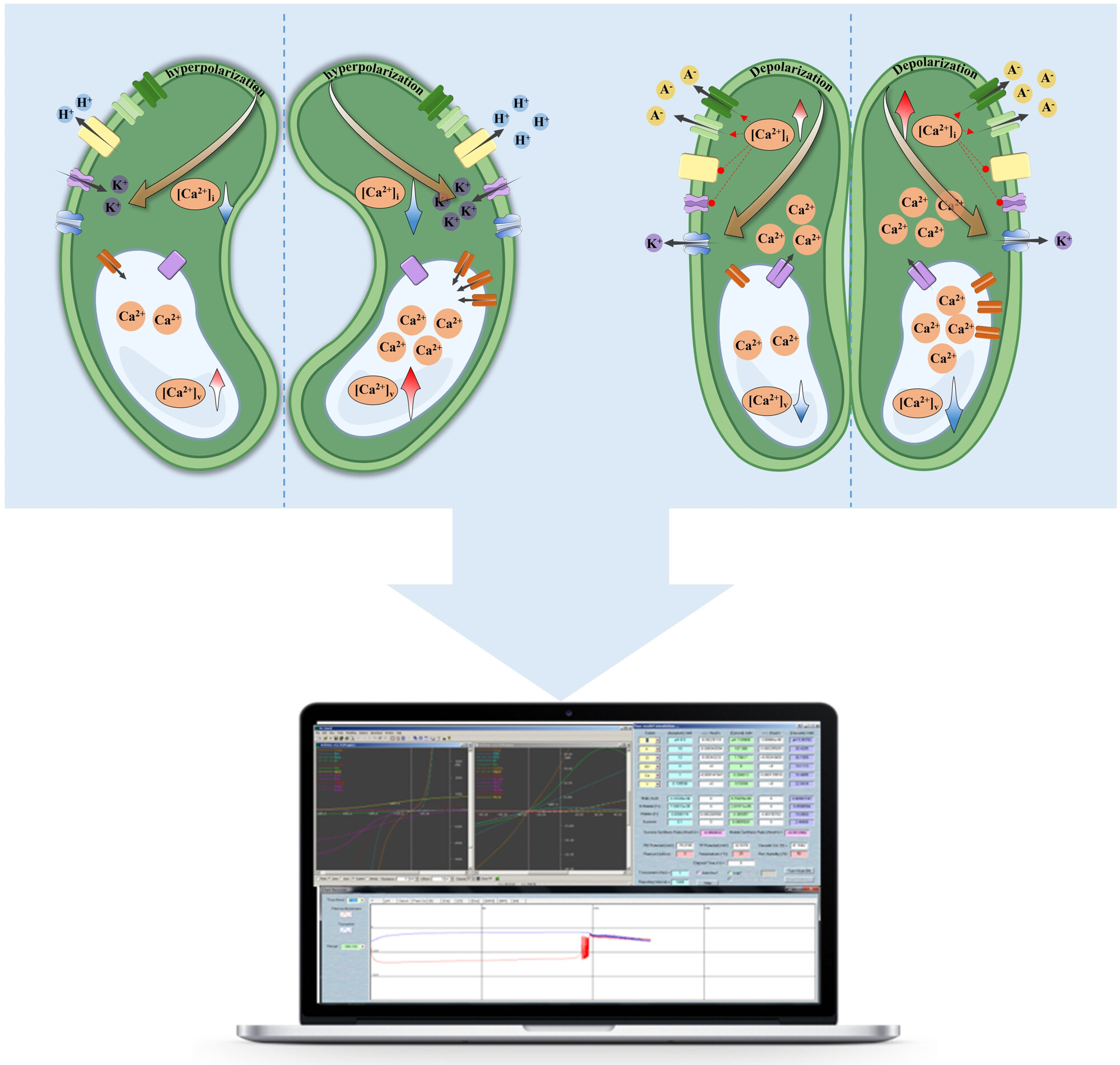
Overexpression of the tonoplast Ca2+-ATPase gene ACA11 increases Ca2+ accumulation in the vacuole, promoting stomatal opening, carbon assimilation, and plant growth under optimal conditions. However, during drought, the over-accumulation of vacuolar Ca2+ triggers a larger release of cytosolic Ca2+, resulting in stomatal closure and enhancing drought tolerance.
Cell and Developmental Biology
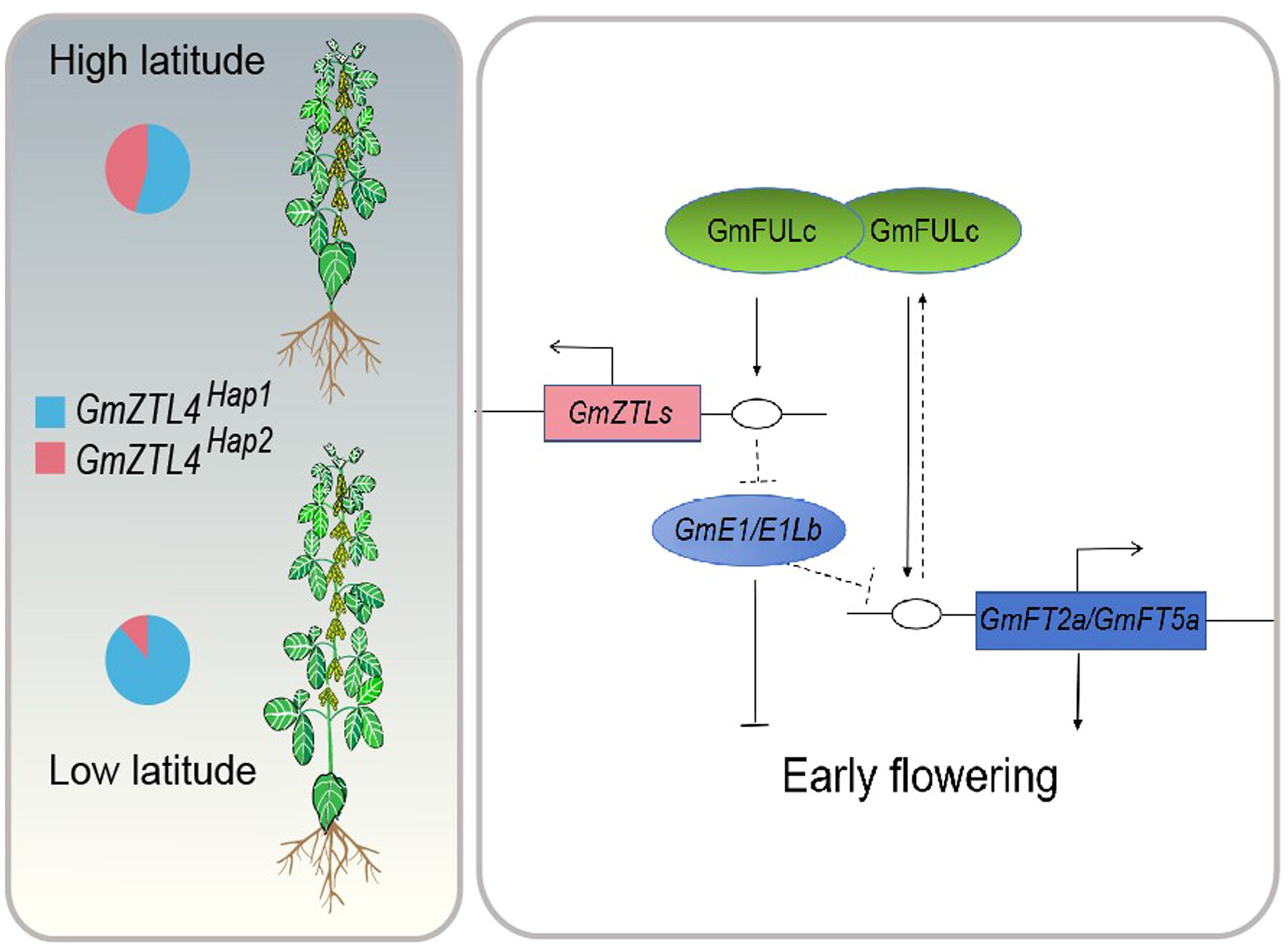
The MADS-box transcription factor GmFULc promotes GmZTL4 gene transcription to modulate maturity in soybean
- Pages: 1603-1619
- First Published: 13 June 2024
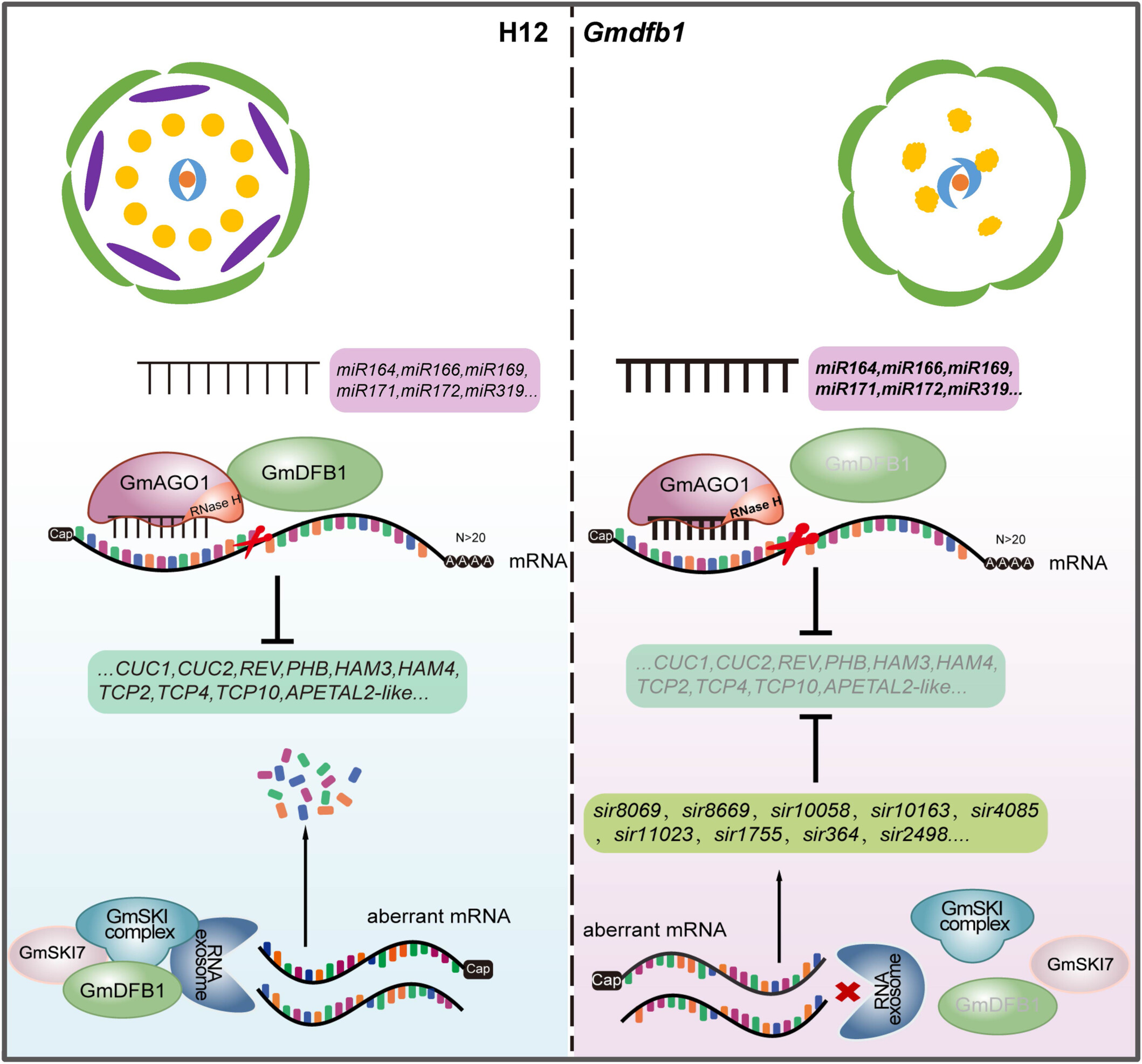
GmDFB1, an ARM-repeat superfamily protein, regulates floral organ identity through repressing siRNA- and miRNA-mediated gene silencing in soybean
- Pages: 1620-1638
- First Published: 11 June 2024
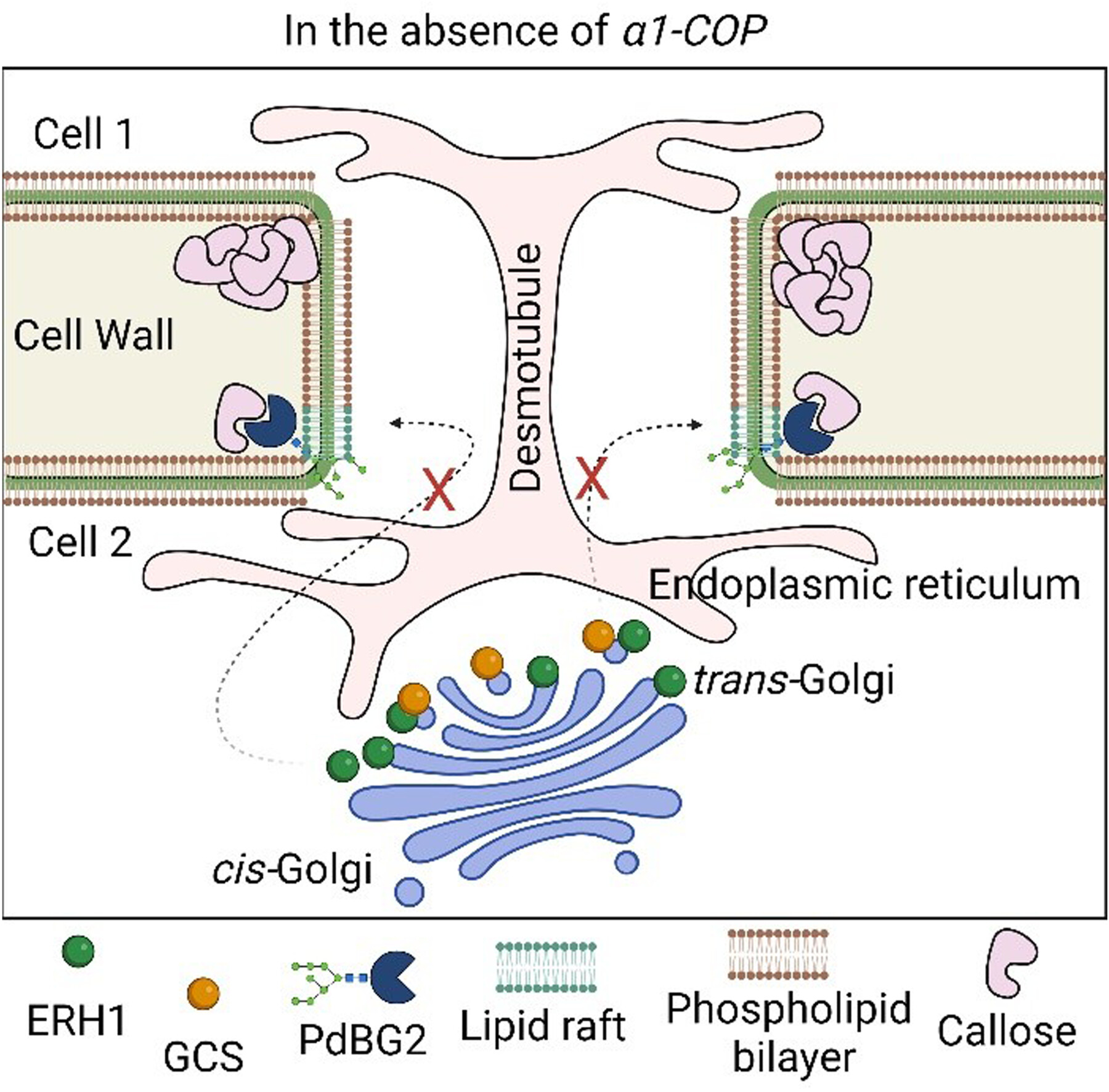
α1-COP modulates plasmodesmata function through sphingolipid enzyme regulation
- Pages: 1639-1657
- First Published: 18 June 2024
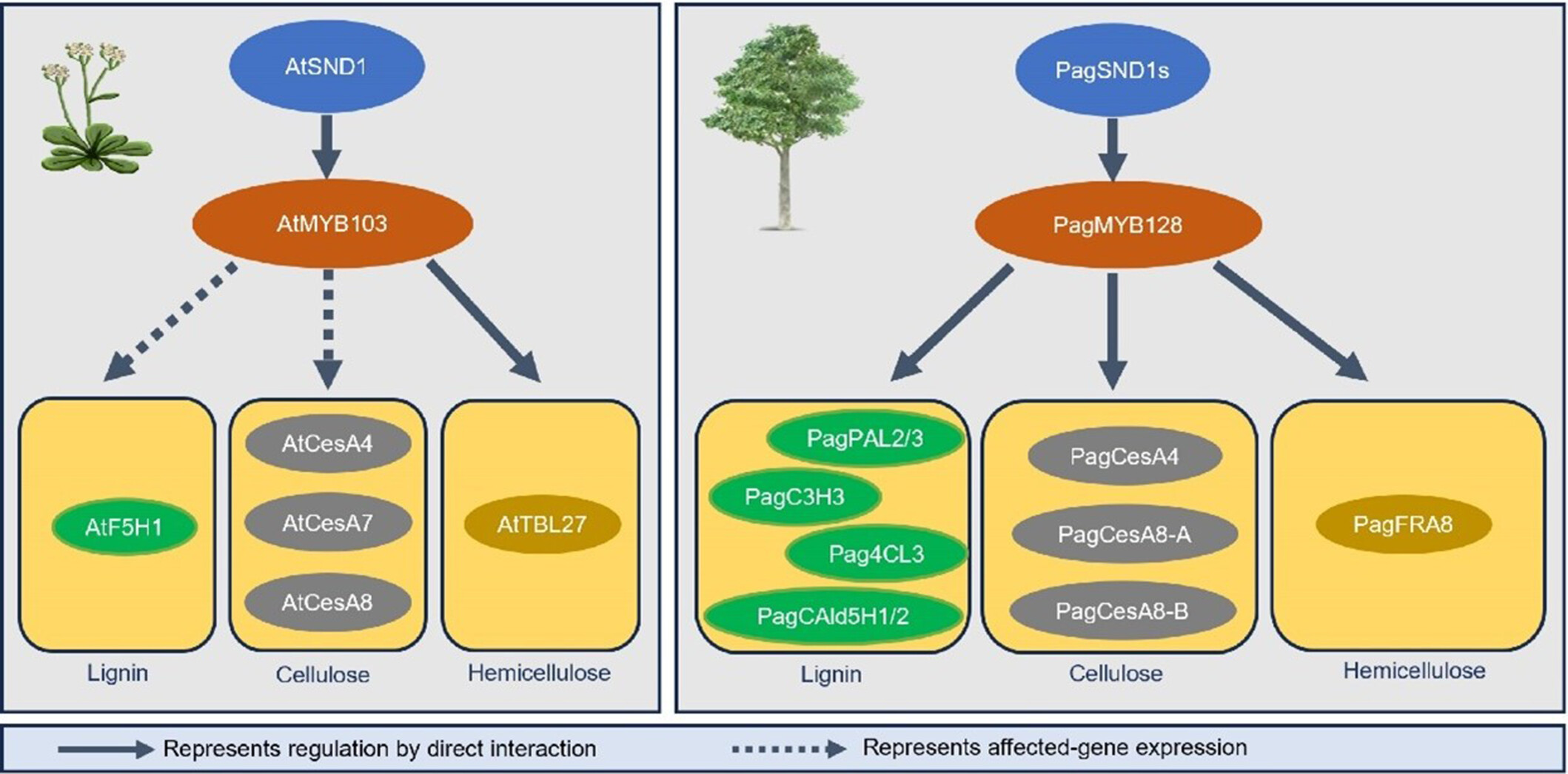
PagMYB128 regulates secondary cell wall formation by direct activation of cell wall biosynthetic genes during wood formation in poplar
- Pages: 1658-1674
- First Published: 21 June 2024
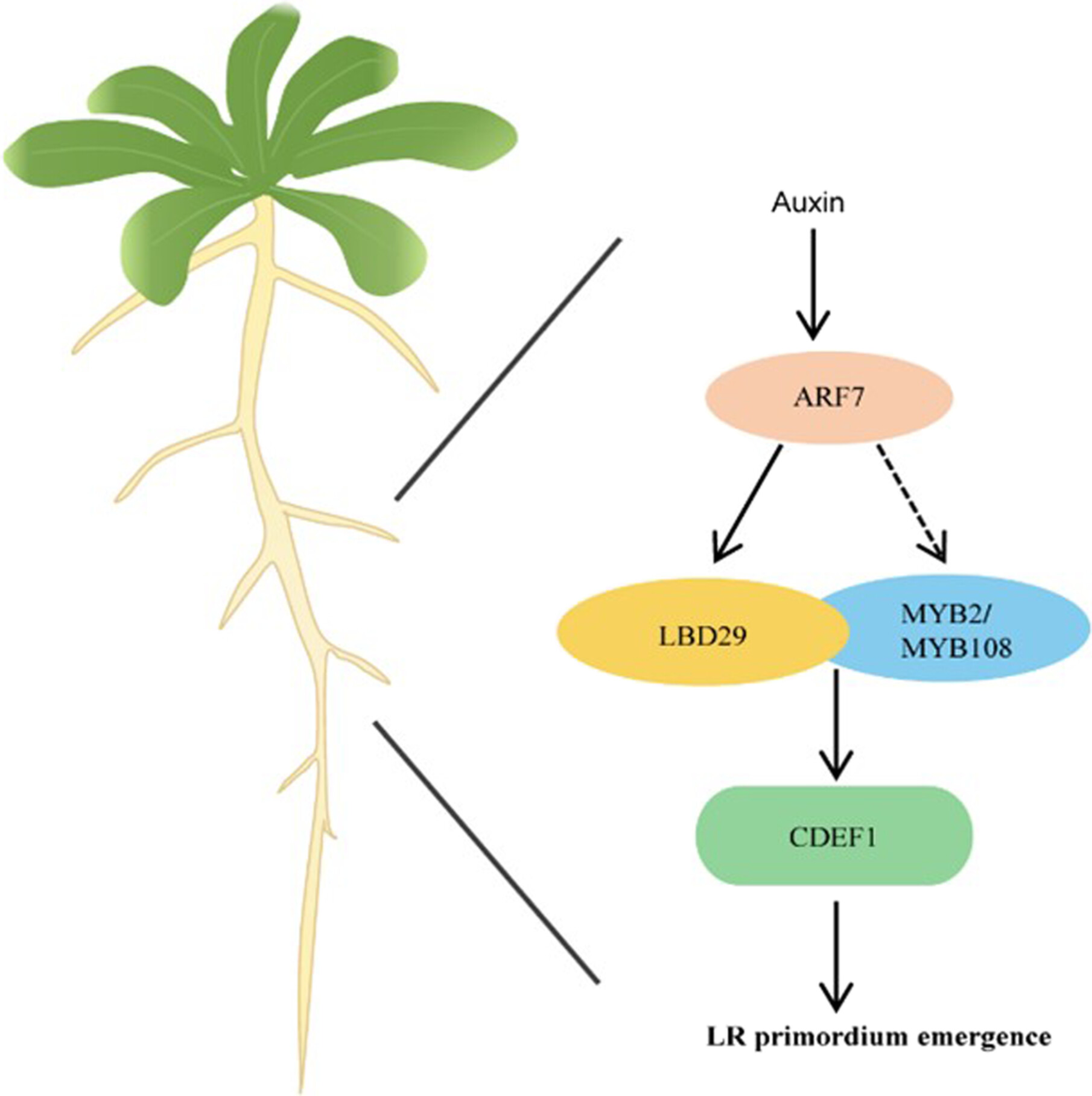
MYB2 and MYB108 regulate lateral root development by interacting with LBD29 in Arabidopsis thaliana
- Pages: 1675-1687
- First Published: 24 June 2024
Functional Omics and Systems Biology
Establishment of genome-editing system and assembly of a near-complete genome in broomcorn millet
- Pages: 1688-1702
- First Published: 02 May 2024
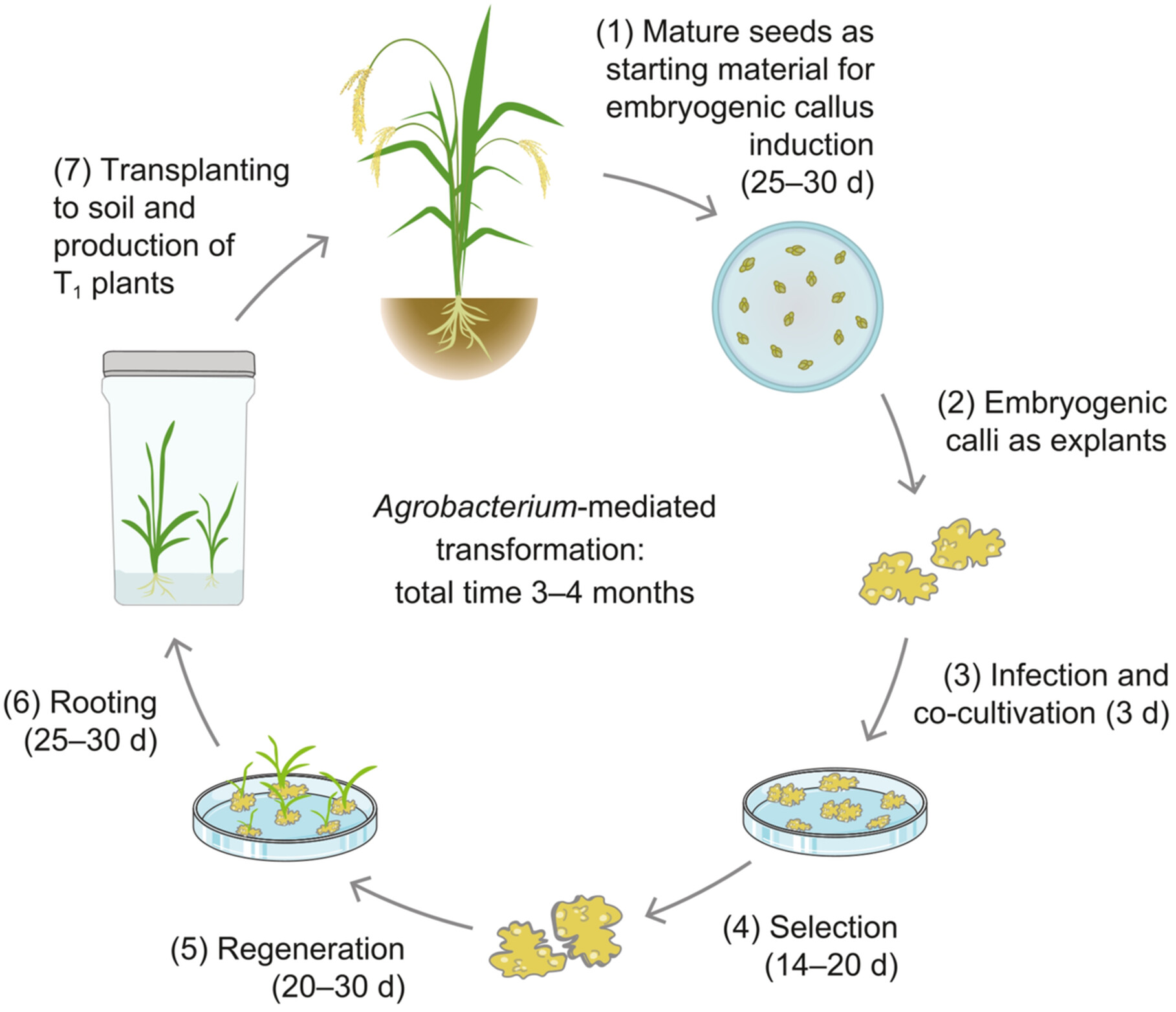
An Agrobacterium tumefaciens-mediated genetic transformation protocol and a CRISPR/Cas9-mediated genome-editing system were established for Hongmi, an elite variety of broomcorn millet (Panicum miliaceum). Additionally, a near-complete, comprehensively annotated genome sequence of Hongmi was assembled. These advances open the door to improving broomcorn millet through biotechnology.
Metabolism and Biochemistry
Identification of the cytochrome P450s responsible for the biosynthesis of two types of aporphine alkaloids and their de novo biosynthesis in yeast
- Pages: 1703-1717
- First Published: 02 July 2024
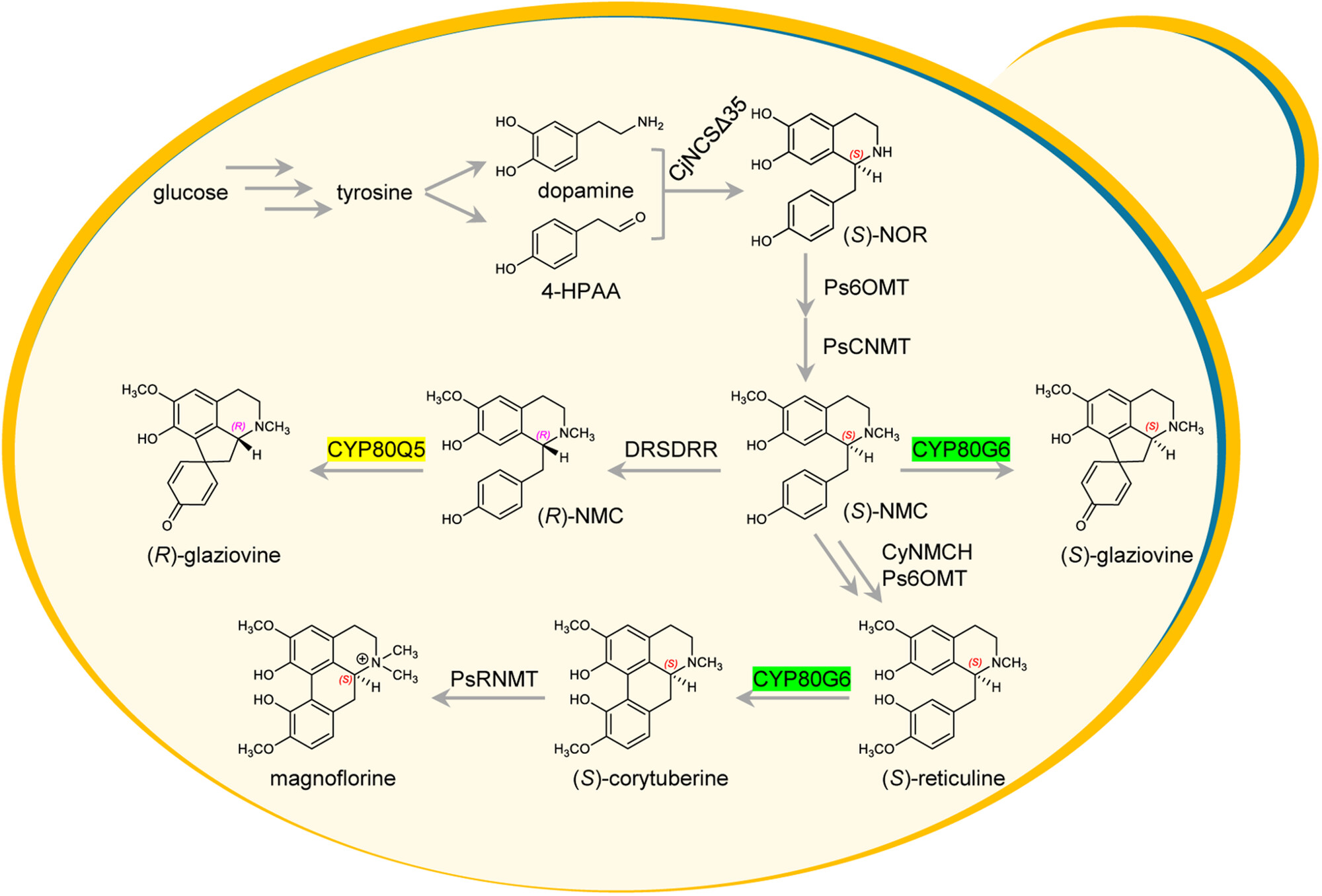
CYP80G6 and CYP80Q5 catalyze the formation of type I and type II aporphine skeletons, respectively. Pathway reconstruction achieved the de novo synthesis of two types of aporphines in yeast. The newly identified CYP719C3 and CYP719C4 are responsible for catalyzing the formation of two methylenedioxy bridges in aporphines.
Molecular Physiology
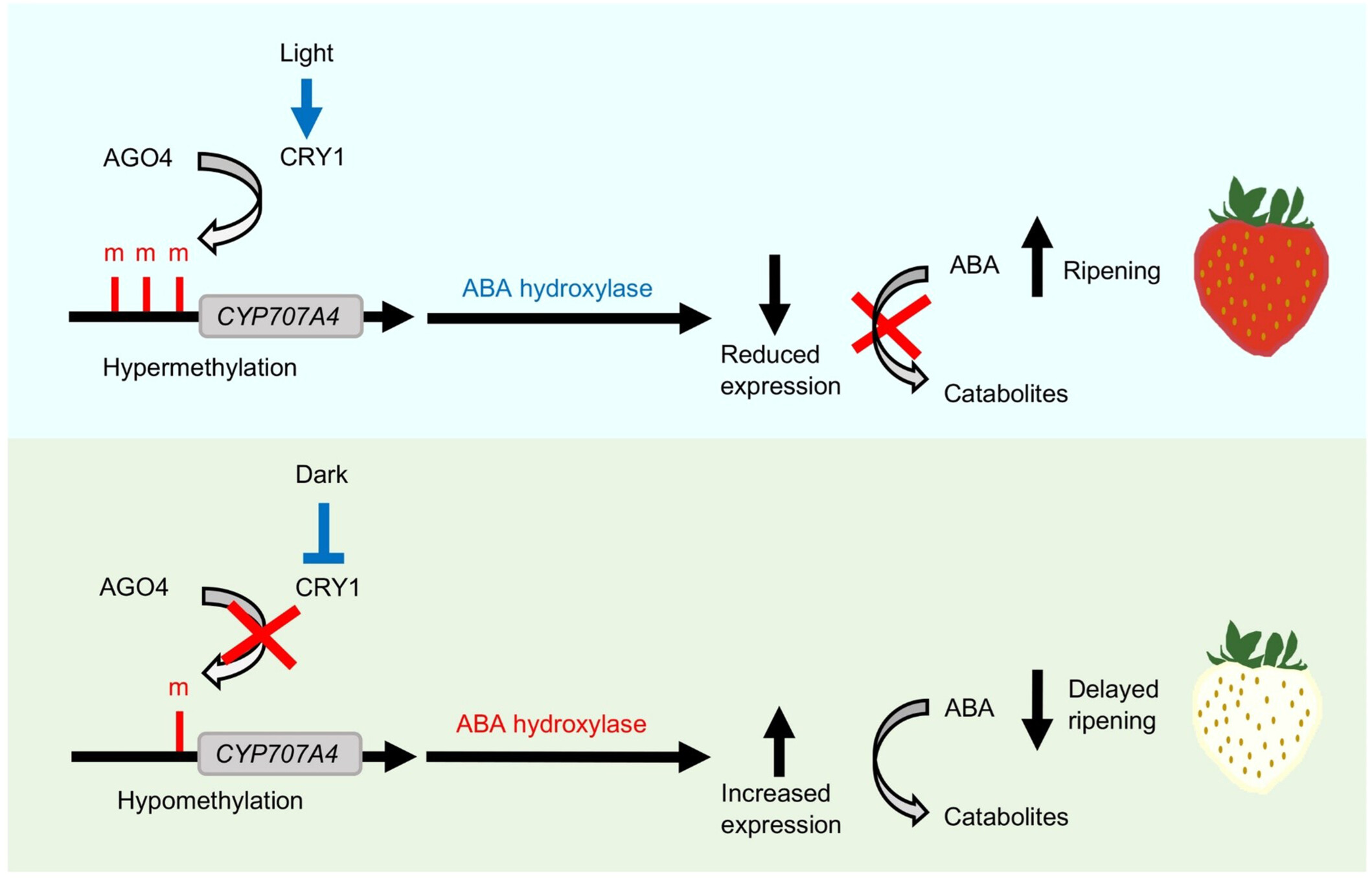
DNA methylation controlling abscisic acid catabolism responds to light to mediate strawberry fruit ripening
- Pages: 1718-1734
- First Published: 19 June 2024
The AaBBX21–AaHY5 module mediates light-regulated artemisinin biosynthesis in Artemisia annua L.
- Pages: 1735-1751
- First Published: 09 July 2024
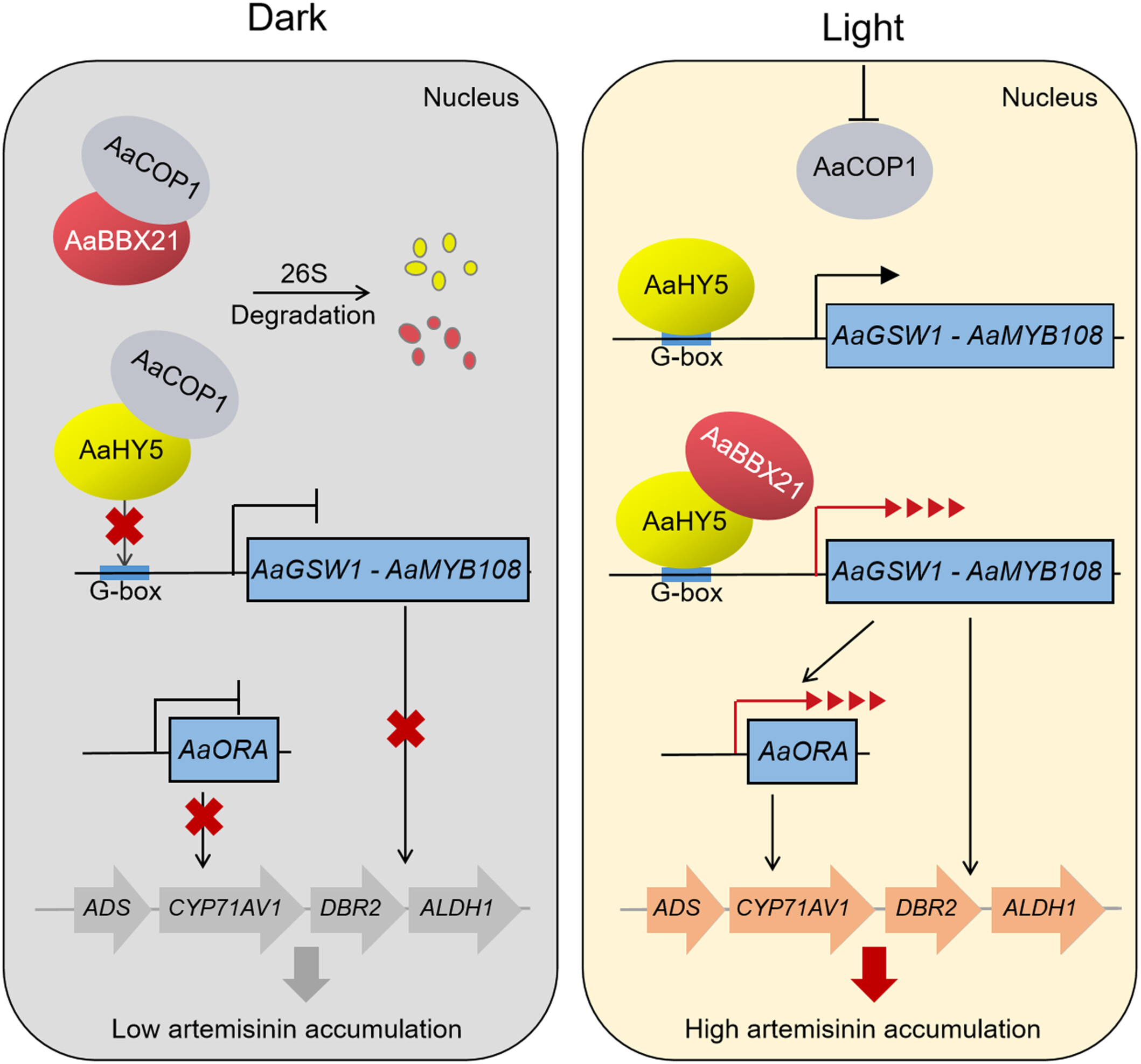
In Artemisia annua, the B-box transcription factor AaBBX21 interacts with ELONGATED HYPOCOTYL5 (AaHY5) to mediate light-regulated artemisinin biosynthesis. The E3 ubiquitin ligase AaCOP1 attenuates AaHY5 and AaBBX21 accumulation, weakening the transcriptional activation activity of the AaBBX21–AaHY5 complex and leading to low artemisinin accumulation in the dark.
A transcriptional cascade involving BBX22 and HY5 finely regulates both plant height and fruit pigmentation in citrus
- Pages: 1752-1768
- First Published: 03 July 2024
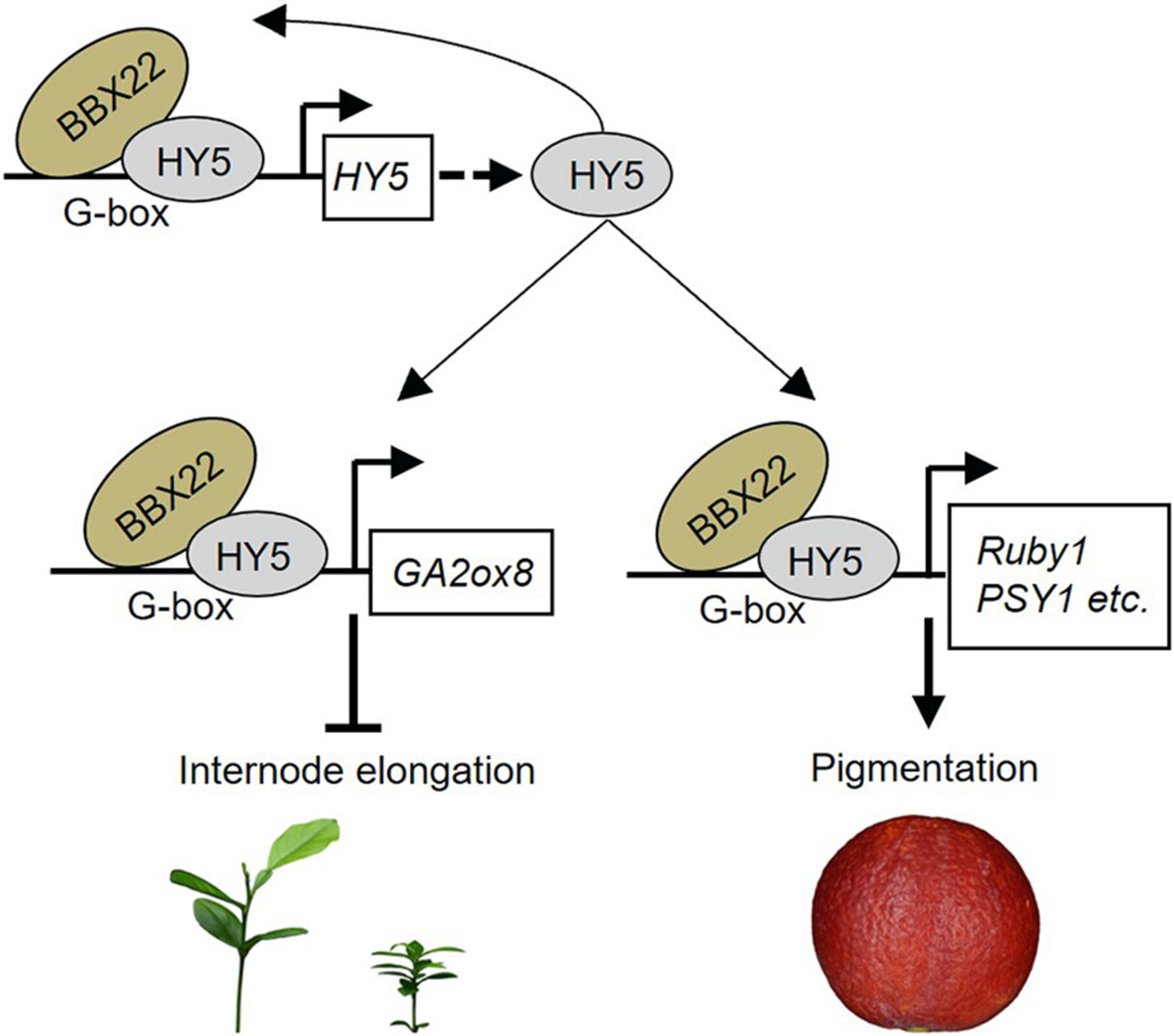
The transcriptional cascade involving B-BOX DOMAIN PROTEIN22 and ELONGATED HYPOCOTYL5 controls internode elongation and participates in the cross-regulation of fruit pigmentation in citrus. Members of this pathway may prove useful for breeding novel types of dwarf trees with attractive, health-beneficial fruits in citrus.
Plant Biotic Interactions
The processed C-terminus of AvrRps4 effector suppresses plant immunity via targeting multiple WRKYs
- Pages: 1769-1787
- First Published: 13 June 2024
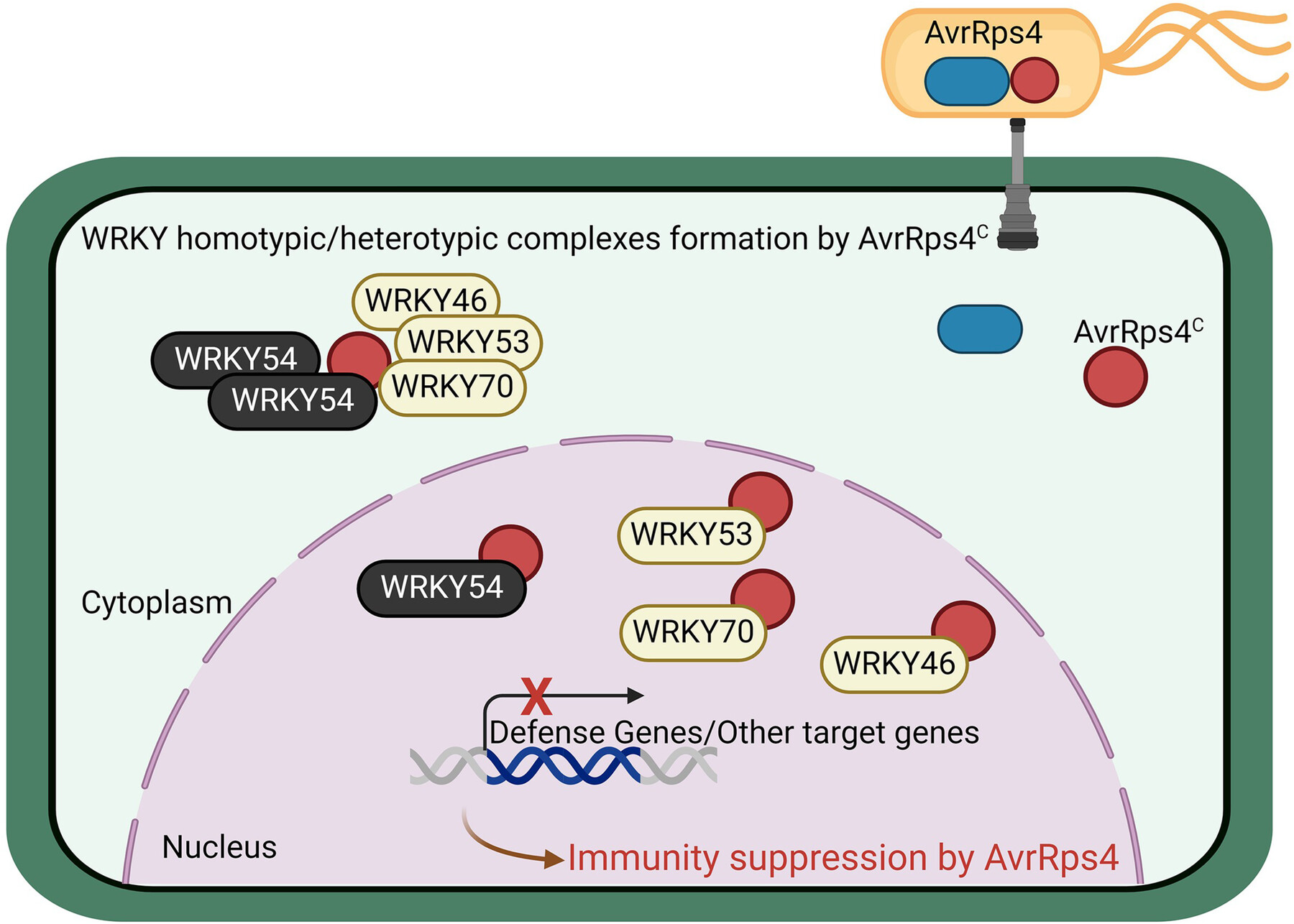
The processed C-terminus of AvrRps4 from Pseudomonas syringae targets the Arabidopsis transcription factors WRKY46, WRKY53, WRKY54, and WRKY70, promoting virulence by suppressing WRKY54-mediated enhanced resistance and WRKY54's transcriptional and binding activities. AvrRps4C induces WRKY-complex formation and translocates WRKYs to the cytoplasm, thus inhibiting WRKY function in plant immunity.
Plant Reproductive Biology
The regulatory mechanism of rapid lignification for timely anther dehiscence
- Pages: 1788-1800
- First Published: 18 June 2024
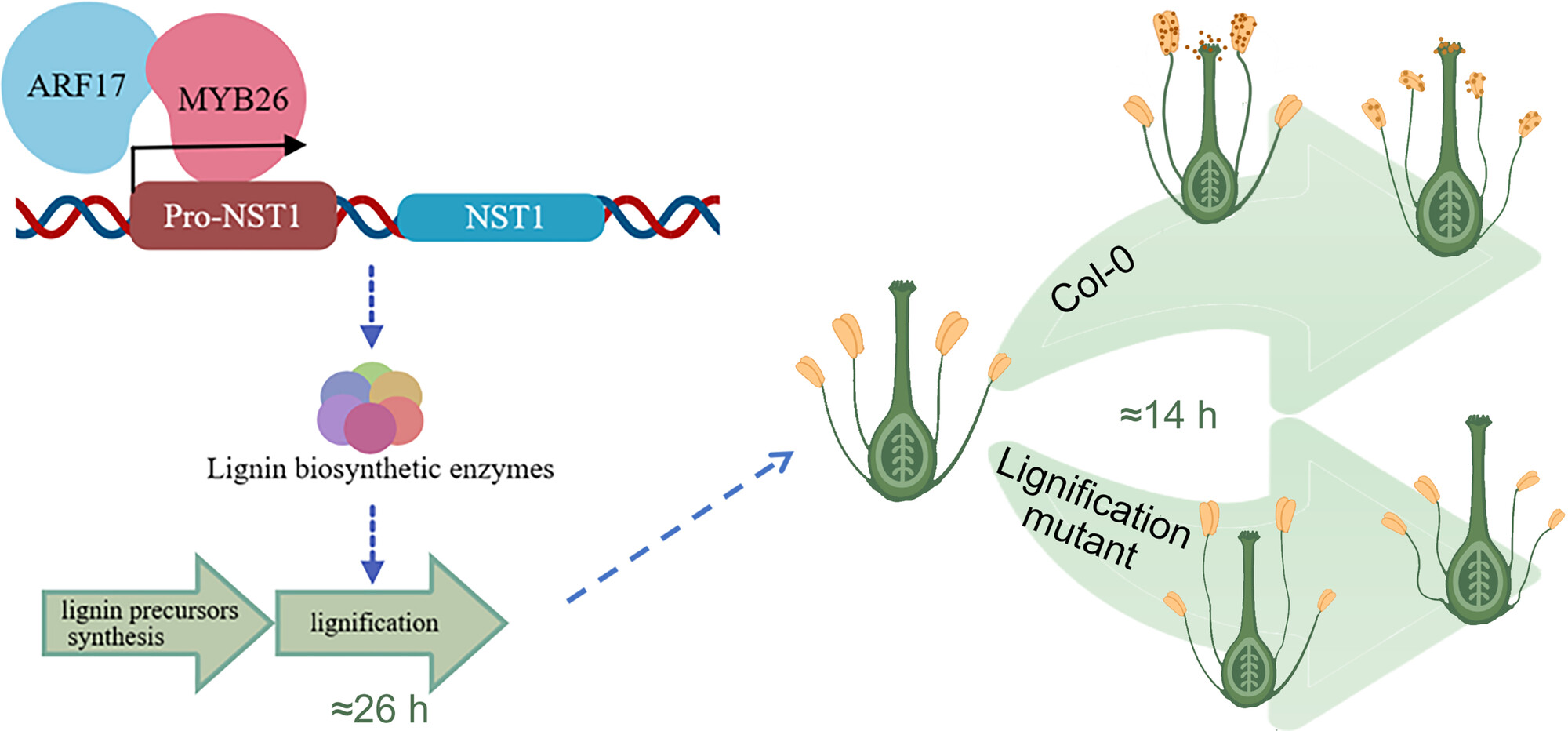
Endothecium lignification can be divided into two phases: the preparation of lignin precursors and the synthesis of lignin. The Arabidopsis AUXIN RESPONSE FACTOR ARF17 acts as a transcription enhancer by interacting with MYB26 to promote expression of the NAC transcription factor genes NST1 and NST2, thereby facilitating endothecium lignification.
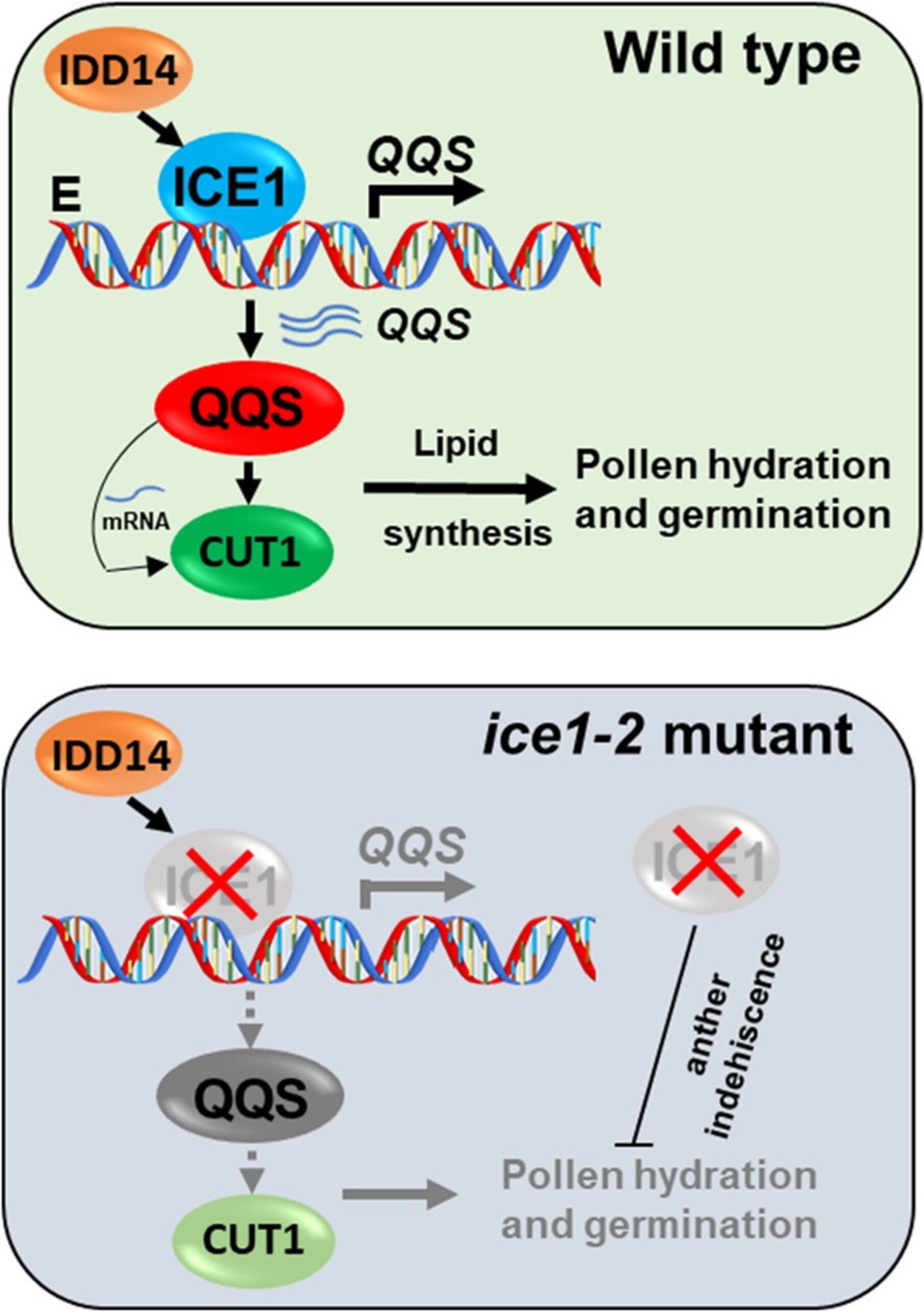
ICE1 interacts with IDD14 to transcriptionally activate QQS to increase pollen germination and viability
- Pages: 1801-1819
- First Published: 28 June 2024