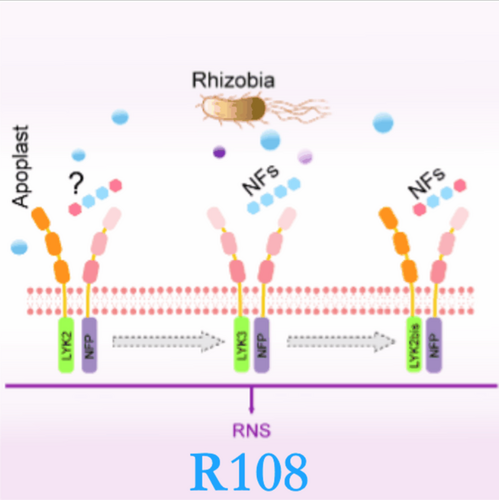
NODULATION TRIO in Medicago truncatula: Unveiling the redundant roles of MtLYK2, MtLYK3, and MtLYK2bis
Edited by: Ertao Wang, Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, CAS, China
Graphical Abstract
The legume–rhizobial symbiosis is a distinctive model in plant–microbe interactions. Despite being foreign invaders, rhizobia are acknowledged as “friends” by compatible host plants, leading to mutualistic endosymbiosis (Cao et al., 2017). Crucial to the establishment of this endosymbiosis is the recognition of rhizobial nodulation factor (NF) by a hetero-receptor complex involving two Lysin Motif receptor kinases (LYKs) in plants, namely NF receptor 1 (LjNFR1), a member in LYK-I clade with kinase activity, and LjNFR5, a member in LYR-I clade without kinase activity (Radutoiu et al., 2003; Madsen et al., 2011). The knockout of either LjNFR1 or LjNFR5 results in a complete loss of rhizobial symbiosis in Lotus japonicus (Radutoiu et al., 2003). While in Medicago truncatula, three LYK genes, including MtLYK3, MtLYK2, and MtLYK2bis, have been identified as orthologs of LjNFR1 (Limpens et al., 2003) (Figure S1). Specifically, M. truncatula cv. Jemalong A17 (hereafter A17) harbors MtLYK2A17 and MtLYK3A17, while M. truncatula spp. tricycla R108 (hereafter R108) possesses MtLYK3R108, MtLYK2R108, and MtLYK2bisR108. Furthermore, it has been pointed out that MtLYK4A17 also plays a role in nodulation (Limpens et al., 2003). However, the expression of MtLYK4A17 failed to complement the nodulation in the A17 lyk3-1 mutant (Luu et al., 2023), suggesting a different function of MtLYK4 in nodulation. In A17, the MtLYK3 knockout mutant, hcl (hair curling), loses the ability to form a shepherd crook and infection thread but keeps some NF-signaling responses, including calcium spiking and expression of some symbiotic genes (Smit et al., 2007). These data supported a two-receptor model in M. truncatula nodulation: MtLYK3 may function as an entry receptor, while the other LYK proteins with uncharacterized functions may serve as signaling receptors (Ardourel et al., 1994; Limpens et al., 2003; Smit et al., 2007). These findings pose an intriguing question of whether these three LYKs coordinate to regulate the rhizobial response. Here, we generated the single, double, and triple mutants in R108 for MtLYK2R108, MtLYK3R108, and MtLYK2bisR108 using CRISPR/Cas technology and examined their roles in nodulation. Our findings suggest that overexpression of any of the three LYK genes can fully mediate nodulation in R108.
In R108, the three LYK genes, MtLYK3R108, MtLYK2R108, and MtLYK2bisR108, are located in the same cluster on chromosome 5 (Figure 1A). This clustered arrangement complicates the generation of double or triple mutants using the traditional cross method. To elucidate the functions of LYKs in nodulation, we designed five guide RNAs (gRNAs) based on conserved DNA regions across the three LYK genes and employed CRISPR/Cas technology to knock out these genes in R108 (Figure S2A). Following multiple rounds of stable transformation assays, we successfully obtained all single, double, and triple mutants for the three LYK genes. Analysis of DNA insertions or deletions in each mutant line revealed frame-shift mutations leading to premature terminations of protein translation. Consequently, all these single, double, and triple mutant plants represent loss-of-function mutations for their respective genes (Figure S2B). To further demonstrate the specificities of all gRNAs and to avoid potential off-target effects, we performed extensive alignments and Sanger sequencing. The results showed that no off-target modifications were detected (Figure S3).
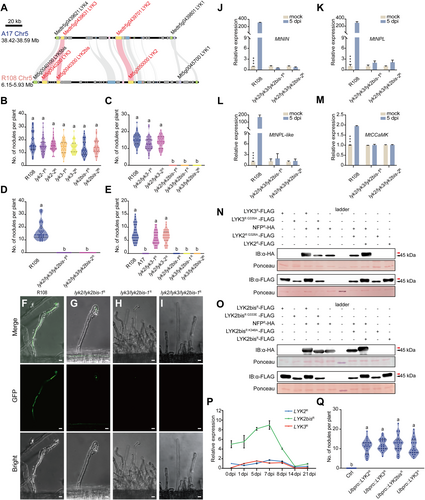
Phenotypic analysis of Medicago truncatula mutants
(A) Microsynteny analysis reveals partial homology between M. truncatula A17 and R108 genomes in the 5th chromosome region. The syntenic blocks encompass specific genes from the LYK-I orthogroup gene family. (B–D) Analysis of nodulation in R108 mutants. Number of nodules at 21 d post-inoculation with Sinorhizobium meliloti 2011 for single mutants (B), double mutants (C), and triple mutants (D). (E) Number of nodules formed on different plants 21 dpi with S. meliloti 2011 nodF/nodL double mutant. Lowercase letters indicate significant differences (n = 30, ANOVA, Tukey, P < 0.05). (F–I) Development of epidermal infection threads in M. truncatula root hairs at 4 dpi. Infection events by S. meliloti 2011 (expressing GFP; green) on WT R108 roots (F), lyk2/lyk2bis-1 mutant roots (G), lyk2/lyk2bis-1 mutant roots (H), and lyk2/lyk2bis/lyk3-1 mutant roots (I). The upper panel shows the merged images, middle panel indicates GFP images, while lower panel shows bright-field images. (Scale bars, 20 μm). (J–M) Expression of symbiosis-related genes in M. truncatula R108 and two triple mutants 5 dpi with S. meliloti 2011 using qPCR. Data are the mean ± SE (n = 3, P < 0.001, one-way ANOVA test). (N, O) MtLYK2R108, MtLYK3R108, and MtLYK2bisR108 exhibit kinase activity, and phosphorylates MtNFPED by inducing band retardation. Phospho-deficient mutations lack phosphorylation activity and are unable to phosphorylate MtNFP. The arrowhead in red indicates band retardation. (P) Relative expression levels of MtLYK2R108, MtLYK3R108, and MtLYK2bisR108 in nodules at various stages post-inoculation, determined using the comparative Ct method, relative to the expression of MtLYK2 in un-inoculated conditions. Data are the mean ± SE (n = 3). (Q) Complementation of nodulation capacity through the overexpression of MtLYK2R108, MtLYK3R108, MtLYK2bisR108, and MtLYK3A17 in lyk2/lyk2bis/lyk3-1 mutants. Recording the nodulation phenotype in transformed roots. Lowercase letters indicate significant differences (n = 20, ANOVA, Tukey, P < 0.05). Gene or mutant followed with the upper letters “R” and “A” represents its origin from R108 and A17, respectively.
To investigate the function of three LYKs in nodulation, we initially assessed nodulation phenotypes in all single mutants: lyk2-1, lyk2-2, lyk3-1, lyk3-2, lyk2bis-1, and lyk2bis-2, 21 d post-inoculation (dpi) with Sinorhizobium meliloti 2011. All single mutant plants exhibited similar nodule numbers compared to the wild-type (WT) plants (Figure 1B). This observation aligns with previous reports indicating no differences in nodule numbers between Tnt1-insertional mutants for each gene and WT plants (Luu et al., 2022). Subsequently, we examined nodulation phenotypes in three double mutant plants. Both the lyk2/lyk2bis and lyk3/lyk2bis double mutants failed to produce any nodules at 21 dpi, contrasting with the lyk2/lyk3 mutant which formed similar nodule numbers as WT plants (Figure 1C). Further analysis of the triple mutant lyk2/lyk3/lyk2bis revealed an inability to form any nodules (Figure 1D).
To gain insights into the inner cell structures of nodules generated on the lyk2/lyk3 double mutant plants, we conducted a toluidine blue staining assay for nodule sections. As shown in Figure S4, the inner cell structures exhibited no observable differences in symbiotic cells between WT and lyk2/lyk3 mutant nodules (Figure S4). These findings collectively underscore the major role of MtLYK2bisR108 in nodulation.
To comprehensively assess nodulation phenotypes, we examined physiological responses, including root hair deformation and infection thread formation, at the early stage of rhizobial infection in these mutants. In the lyk2/lyk2bis and lyk3/lyk2bis mutant plants, 4 dpi with GFP-labeled S. meliloti 2011, we observed root hair deformation and the presence of infection foci, but not the formation of infection threads (Figure 1G, H), as compared to WT plants (Figure 1F). However, in the lyk2/lyk3/lyk2bis triple mutant plants, we could hardly observe any root hair deformation, infection foci, or infection threads (Figure 1I). To confirm and quantify the NF responsiveness of the mutants, we studied hair curling responses to purified NF in both the lyk2/lyk3/lyk2bis triple mutant and various double mutants. Following treatment with 10−8 M NF, hair curling in the triple mutant was essentially unobservable compared to the WT plants (Figure S5A). In contrast, NF treatment of the double mutants revealed distinct differences in hair curling responses. The lyk2/lyk2bis and lyk3/lyk2bis double mutants that failed to form nodules showed significantly fewer hair curling events compared to the WT, whereas the lyk2/lyk3, exhibited hair curling numbers similar to the WT plants (Figure S5B). Furthermore, we observed a complete block in the transcriptional induction of some symbiosis-related genes, including Nodule Inception (NIN), NIN-Like (NPL), NPL-like, and Calcium/Calmodulin-Dependent Protein Kinase (CCaMK) (Roy et al., 2019), in the lyk2/lyk3/lyk2bis triple mutants compared to the high expression levels in WT nodules (Figure 1J–M). These data indicate that the lyk2/lyk3/lyk2bis triple mutants completely lose the response to rhizobium, mirroring observations in L. japonicus nfr1 mutants. While both double mutant plants, lyk3/lyk2bis and lyk2/lyk2bis, failed to produce nodules, they still retained responses to rhizobium attachment, albeit without establishing effective rhizobial invasion. These findings suggest that MtLYK2R108 and MtLYK3R108 might have redundant roles in mediating rhizobium recognition and/or attachment.
Previous reports have confirmed MtLYK2bisR108 extends the nodulation specificity of R108 to the S. meliloti nodF/nodL mutant strain (Luu et al., 2022). We separately inoculated three double mutants with nodF/nodL mutant strain and found that only lyk2/lyk3-1 and lyk2/lyk3-2 could generate a similar number of nodules as WT plants (Figure 1E). This indicates that MtLYK2bisR108 might simultaneously function as both an entry receptor and a signaling receptor for recognizing both nodF/nodL mutant strain and WT S. meliloti.
The above data suggest that all three receptors, MtLYK2R108, MtLYK3R108, and MtLYK2bisR108, may possess redundant functions in mediating symbiotic signaling transduction. An intriguing observation arises from the co-expression of LjNFR1/MtLYK3 and LjNFR5/MtNFP (NF perception in M. truncatula, the ortholog of LjNFR5), which induces cell death in Nicotiana benthamiana leaves (Madsen et al., 2011; Pietraszewska-Bogiel et al., 2013). To probe the associations between MtLYK2R108, MtLYK3R108, and MtLYK2bisR108 and MtNFP, we co-overexpressed MtNFP with either MtLYK2R108, MtLYK3R108, or MtLYK2bisR108 in N. benthamiana. Remarkably, we observed apparent cell death in leaves upon co-overexpressing MtNFP with any of the receptors, compared to the control (Figure S6).
To investigate the potential involvement of MtLYK2R108, MtLYK3R108, and MtLYK2bisR108 in symbiotic signaling, extracellular domain (ED) structures of these receptors were predicted using the known structure of A17 MtLYK3ED as a template in SwissModel (Figure S7). The results revealed a high degree of similarity in three-dimensional structures among the three receptors.
Given the established transphosphorylation between LjNFR1/MtLYK3 and LjNFR5/MtNFP in transducing NF-signaling in plant cells (Madsen et al., 2011; Luu et al., 2022, 2023), we investigated whether MtLYK2R108, MtLYK3R108, and MtLYK2bisR108 share similar functions in phosphorylating MtNFP. Co-expression experiments involving the cytoplasmic domains (CDs) of MtLYK2R108, MtLYK3R108, and MtLYK2bisR108, along with their kinase-dead versions, and the CD of MtNFP or controls were conducted in Escherichia coli cells. The results revealed an apparent band retardation of MtNFPCD in the presence of either MtLYK3CD, MtLYK2CD, or MtLYK2bisCD, but not their kinase-dead versions (Figure 1N, O). Immunoblots further identified the retardation of bands for MtLYK2CD, MtLYK3CD, and MtLYK2bisCD, indicating their capacity for kinase activity with auto-transphosphorylation, as well as trans-phosphorylation of MtNFP.
The above data suggest that MtLYK2R108, MtLYK3R108, and MtLYK2bisR108 exhibit redundant functions, with MtLYK2bisR108 playing a major role in mediating nodulation in M. truncatula. This observation prompted us to speculate about the differential regulation of transcription levels for MtLYK2R108, MtLYK3R108, and MtLYK2bisR108. To explore this speculation, we examined the transcriptional levels of three LYK genes in M. truncatula roots and nodules at different time points post-inoculation with rhizobium. Both MtLYK2R108 and MtLYK3R108 displayed low expression levels in the roots before and after rhizobial treatment. In contrast, the expression of MtLYK2bisR108 was higher than that of MtLYK2R108 and MtLYK3R108 (Figures 1P, S8). The elevated expression levels of MtLYK2bisR108 may contribute to its prominent role in mediating nodulation in M. truncatula.
To validate the hypothesis that all three receptors have redundant functions, we conducted complementation assays by expressing MtLYK2R108, MtLYK3R108, and MtLYK2bisR108 driven by ubiquitin promoter in the lyk2/lyk3/lyk2bis-1 triple mutant plants. Intriguingly, the overexpression of any of MtLYK2R108, MtLYK3R108, or MtLYK2bisR108 completely restored nodulation in the lyk2/lyk3/lyk2bis mutant plants. Similar numbers of nodules were formed on each transgenic root compared with the control plants (Figures 1Q, S9A–E). These data strongly suggest that all three LYKs shared a similar function in mediating nodulation in M. truncatula.
In conclusion, our comprehensive study on multiple M. truncatula mutant plants has revealed that all three receptors, MtLYK2R108, MtLYK3R108, and MtLYK2bisR108, exhibit redundant functions in mediating nodulation. The overexpression of any of these receptors could complement nodulation defects in the lyk2/lyk3/lyk2bis mutant plants. Notably, among these receptors, MtLYK2bisR108 appeared to play a major role in nodulation, probably due to its relatively high expression levels compared with the other two genes in response to rhizobial inoculation. The differential expression patterns of the three LYK genes may be crucial for the precise regulation of nodulation in M. truncatula, although the underlying mechanisms are yet to be determined. These observations, along with previous publications challenging the two-receptor model in rhizobial symbiosis, suggest that a one-receptor model with multiple specificities that mediates both signaling and entry responses may be more suitable in M. truncatula. These data advance our understanding of nodulation processes and provide valuable genetic materials for studying rhizobium symbiosis.
ACKNOWLEDGEMENTS
We extend our sincere gratitude to Professors Christian Staehelin, Erik Limpens, and Eva Kondorosi for kindly providing the rhizobium strains. This work was supported by the National Key R&D Program of China (2019YFA0904700), the National Natural Science Foundation of China (32090063), and a Self-Innovation grant from the National Laboratory (AML2023B01). The qPCR and microscopy data are acquired from the Core Facility Center run by the National Lab of Agricultural Microbiology.
CONFLICTS OF INTEREST
The authors declare no competing interests.
AUTHOR CONTRIBUTIONS
Y.C. and Y.L. conceived the project. Y.L., Y.Z., and Z.Y. performed all the experiments. R.D. and H.Y. provided substantial help in experiments. H.Y. and H.Z. provided valuable suggestions for the study. Y.L., H.Z., and Y.C. analyzed the data and finalized all figures with inputs and comments from co-authors. Y.L. and Y.C. wrote the article. All authors read and approved of the manuscript.