Developing guanine base editors for G-to-T editing in rice
Edited by: Dr. Lanqin Xia, Institute of Crop Sciences, CAAS, China
Graphical Abstract
Two guanine base editors created using an engineered N-methylpurine DNA glycosylase with CRISPR systems achieved targeted G-to-T editing with 4.94–12.50% efficiency in rice (Oryza sativa). The combined use of the DNA glycosylase and deaminases enabled co-editing of target guanines with adenines or cytosines.
Single nucleotide polymorphisms (SNPs) are widely present and related to desirable agronomic traits in crops. clustered regularly interspaced palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated base editors have been frequently used to correct defective alleles and create novel alleles by artificial evolution for rapid crop genetic improvement (Ma et al., 2021). In the past several years, the cytosine and adenine base editors (CBEs and ABEs) (Ren et al., 2018; Yan et al., 2021) have been extensively developed and are widely utilized for C-to-T and A-to-G conversions in plants, respectively. Also, their derivative C-to-G base editors (CGBEs) and adenine transversion base editor (AKBE) were successfully established (Koblan et al., 2021; Wu et al., 2023). These base editing methods all involve the deamination of cytosine (C) or adenine (A) residues as the first crucial step, resulting in the formation of deoxyuridine (U) or deoxyinositol (I) intermediates, which are subsequently converted into other bases (Tong et al., 2023b). Very lately, a base editor named glycosylase-based GBE (gGBE) achieved efficient G-to-Y (Y = C or T) editing in mammal cells by using N-methylpurine DNA glycosylase (MPGv6.3) and SpCas9n (Tong et al., 2023a). MPGv6.3 is able to glycosylate the target G and then the base excision repair (BER) pathway is initiated to remove the glycosylated G and generate the apurinic/apyrimidinic (AP) sites, finally resulting in G-to-Y base editing via translesion synthesis (TLS) or DNA replication. However, whether DNA glycosylase can be employed to develop GBE and derivative base editors for generating nucleotide changes in plants remains elusive.
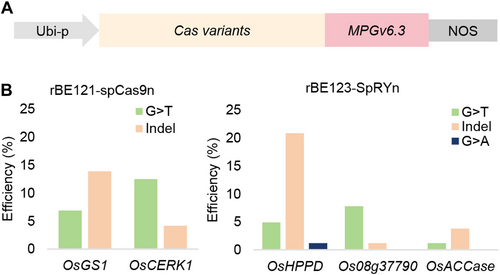
To explore the potential of G-to-Y editing in the rice genome, the engineered glycosylase variant MPGv6.3 was optimized using rice codons and fused to the 3′-terminal of SpCas9n, resulting in rBE121 (Figure 1A). We designed two single-guide RNAs (sgRNAs) to target endogenous OsCERK1 and OsGS1 toward the canonical NGG protospacer adjacent motifs (PAMs), respectively. After Agrobacterium-mediated transformation of rice callus, independent transgenic lines were genotyped by directly sequencing the polymerase chain reaction (PCR) amplicons of the target regions, we found that only G-to-T transversion, no G-to-C editing, were generated at the OsCERK1 and OsGS1 target sites, with editing efficiency of 12.50% (6/48) and 6.97% (3/43), respectively (Figures 1B, S1). Of note, the occurrence of indel (insertion and/or deletion) mutations is concomitant with this process (4.17% at OsCERK1 and 13.95% at OsGS1) (Figures 1B, S1). These findings suggest that the DNA glycosylase MPGv6.3 is functional in rice and can be employed in base editing, but the efficiency of MPGv6.3-mediated GBE editing is currently suboptimal. The editing efficiency of GBE can be enhanced by searching for MPG homologs such as mouse MPG and engineering them, and the substrate specificity and glycosylase activity of MPGv6.3 can also be improved by structure-guided consistent engineering.
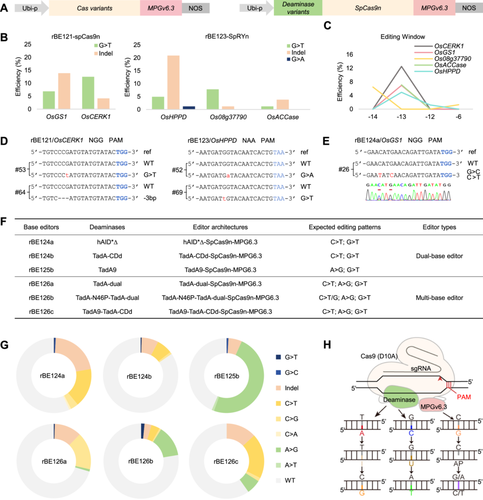
The editing events generated by guanine base editors (GBE) and derivative base editors at various loci in rice genome
(A) Schematic diagrams of GBE constructs (left) and derivative BE constructs (right) for rice base editing. (B) Sequencing results of the rBE121- and rBE123-induced target DNA mutations in T0 transgenic rice plants. (C) The activity windows of rBE121 and rBE123 in rice. (D) Representative mutation types of OsCERK1 and OsHPPD induced by rBE121 and rBE123. The protospacer adjacent motifs (PAM) and the base editing events are marked in blue and red, respectively. (E) Representative Sanger sequencing chromatograms of the OsGS1 target site showing desired C-to-T and G-to-C editing events. The nucleotide mutations in the sequencing chromatograms are highlighted by a red line, and the PAM is marked in blue. (F) Information on derivative BEs. (G) Types and percentages of different editing pattern induced by derivative BEs. (H) Schematic diagram of the working model of the derivative BEs. An nCas9-sgRNA complex creates an R-loop at the target site in the DNA. Deaminases, including cytosine deaminases, adenine deaminases and their dimers, can achieve base editing of A or C through deamination. The glycosylase MPGv6.3 can remove G, and the apurinic/apyrimidinic (AP) site generated is repaired by DNA replication, leading to G-to-C or G-to-T conversion. When the deaminases and the glycosylase co-exist, they occupy the common edit window, theoretically allowing for co-editing A or C and G within the editing window.
Next, we updated pUbi:rBE121 by replacing SpCas9n with SpRYn (Xu et al., 2021) to expand the targeting scope of GBE in rice, resulting in pUbi:rBE123. Three sgRNAs were designed to target OsACCase, Os08g37790, and OsHPPD toward different PAMs, respectively. As shown in Figures 1B and S1, the G-to-T editing efficiency was 7.14% (3/42) at OsACCase, 7.79% (6/77) at Os08g37790, and 4.94% (4/81) at OsHPPD; of note, we observed a G-to-A editing in OsHPPD in this case, with a low efficiency of 1.23% (1/81). Meanwhile, relatively high frequencies of indel mutations were also observed in the target regions (Figures 1B, D, S1). By analyzing all data above, we found that the activity window of MPGv6.3-mediated rice GBE was approximately 9 bp (spanning from protospacer positions 6–14) (Figure 1C). Combined, these data suggest that MPGv6.3 enables dominant G-to-T transversion in rice base editing.
Considering the above-mentioned facts, we attempted to develop potential derivative base editors through combined use of the DNA glycosylase MPGv6.3 with various nucleotide deaminases (Figure 1A). Cytosine deaminases hAID*∆ and TadA-CDd (Chen et al., 2023) as well as adenine deaminase TadA9 were fused to the N-terminal of rBE121, resulting in dual-base editors rBE124a, rBE124b, and rBE125b, respectively (Figures 1A, F, S2). Meanwhile, TadA-dual, an evolved deaminase protein which enables both A-to-G and C-to-T conversion in mammalian cells (Chen et al., 2023), was fused to the N-terminal of rBE121 as well, resulting in multi-base editor rBE126a (Figures 1A, F, S2). Furthermore, TadA dimer strategy, which included TadA-N46P (Chen et al., 2023) and TadA-dual dimer as well as TadA9 and TadA-CDd dimer, was used to construct other multi-base editors rBE126b and rBE126c (Figures 1A, F, S2).
The performance of these derivative base editors was evaluated by detecting sequence diversification of the NGG PAM target regions in independent transgenic rice lines and we observed co-editing events at three target loci (OsGS1, OsALS1, and OsALS1-3). Among them, rBE124a caused C-to-T and G-to-C co-editing at OsGS1 with a frequency of 0.89% (Figures 1E, S3), rBE125b caused A-to-G and G-to-T co-editing (2.41% frequency) as well as A-to-G and G-to-C co-editing (1.04% efficiency) at OsALS1 and OsALS1-3, respectively (Figure S3). Other than that, all these base editors mainly caused nucleotide substitutions of C or A at each target site, rarely functioned on G (Figures 1G, S2), suggesting that most of the editing events were from deaminase activity. Therefore, more nucleotide deaminase variants or long peptide linkers would be recommended in further optimization of derivative base editors containing glycosylase. Anyhow, we constructed a schematic diagram illustrating the operational principles of the derivative base editor (Figure 1H).
In summary, we have successfully developed rice GBEs which achieve dominant G-to-T transversion and have demonstrated the potential of combined use of DNA glycosylase and deaminases in developing derivative base editors for rice genome editing. These constructs are new additions to the current genome-editing toolkit for both basic research and genetic improvement of rice.
ACKNOWLEDGEMENTS
The project was supported by the Biological Breeding-Major Projects (2023ZD04074), the Nanfan special project of the Chinese Academy of Agricultural Sciences (YBXM2313) and the Hainan Seed Industry Laboratory (B23CJ0208) and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences.
CONFLICTS OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
H.Z., X.Z., and L.L. designed the experiments; L.L. and Z.Z. performed experiments; C.W. and F.Y. carried out bioinformatics analysis; H.Z, W.M., and W.S. supervised the experiments; H.Z., L.L., and Z.Z. wrote the paper with input from all other authors. All authors read and approved the contents of this paper.