Journal list menu
Export Citations
Download PDFs
ISSUE INFORMATION
ORIGINAL ARTICLES
High-volume plankton tow net sampling improves eDNA detection of invasive zebra mussels (Dreissena polymorpha) in recently infested lakes
- First Published: 12 March 2024
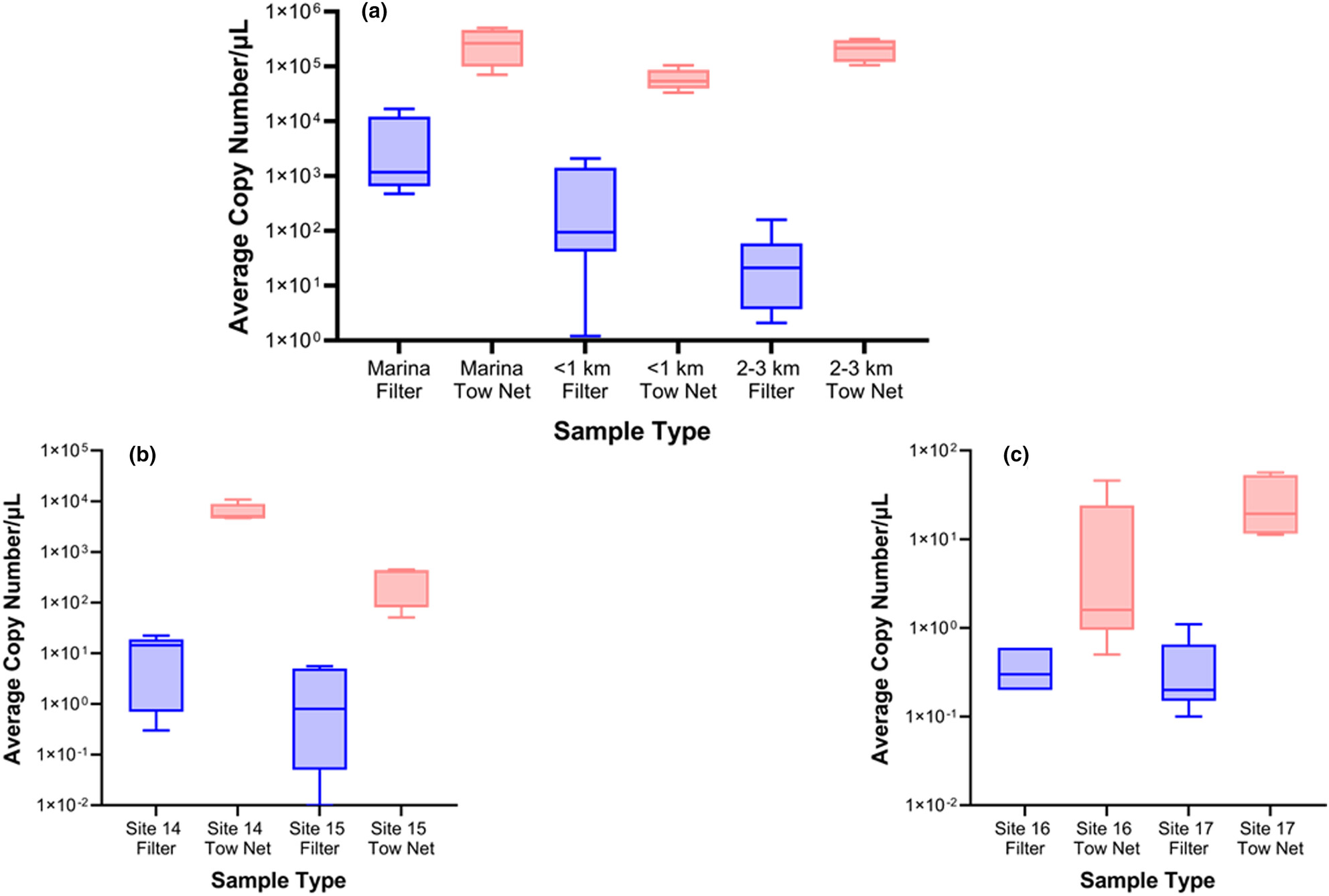
Environmental DNA (eDNA) techniques offer sensitive monitoring tools for the early detection of aquatic invasive species, which is increasingly needed to prevent their establishment and spread. We compared the eDNA sensitivities of high-volume plankton tow net water sampling to conventional low-volume water filtration methods in three lakes of known recent zebra mussel (Dreissena polymorpha) infestations. Paired filtration and tow net samples were analyzed for Dreissena DNA where higher yields of Dreissena eDNA (more DNA copies) were recovered from plankton tow than from filtered samples in all 33 paired comparisons.
PERSPECTIVE
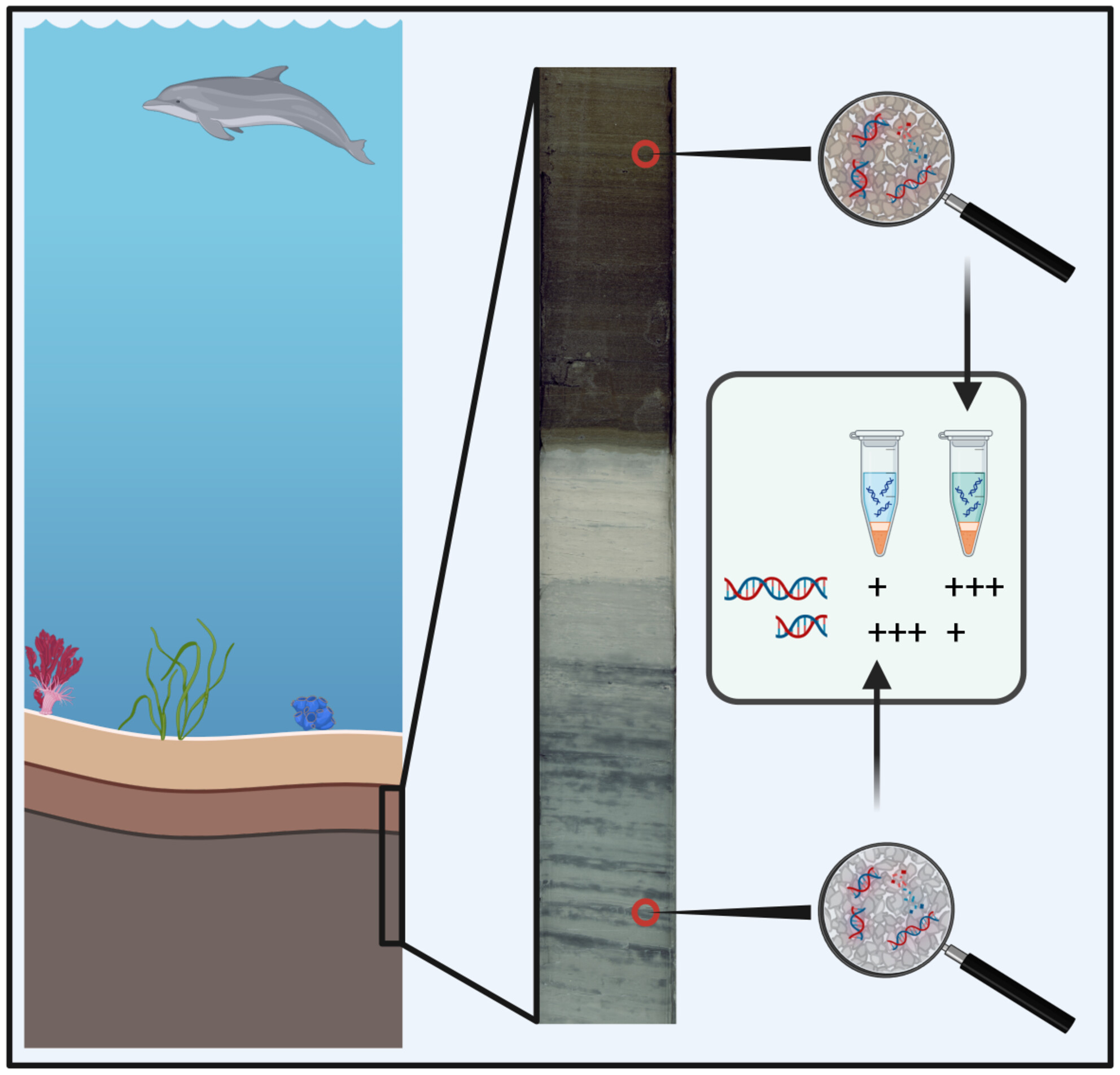
Importance of eDNA taphonomy and sediment provenance for robust ecological inference: Insights from interfacial geochemistry
- First Published: 11 March 2024
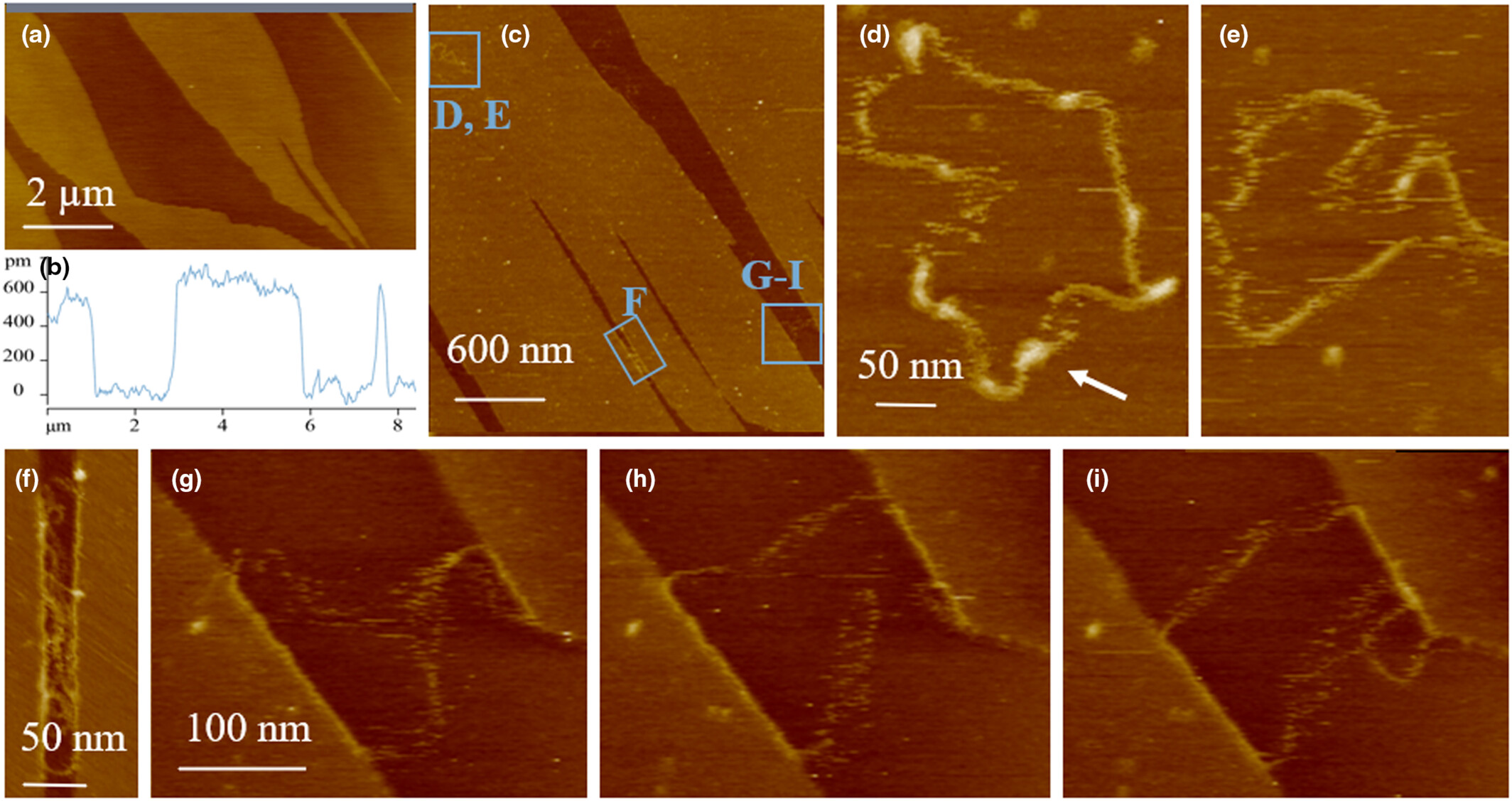
We tune in on the importance of understanding eDNA taphonomy and sediment provenance for the field of eDNA research. We use atomic force microscopy to show how interfacial geochemical interactions drive DNA adsorption behavior on mineral surfaces and combine mineralogic composition with experimental adsorption data to outline a way forward to increase the scope and resolution of biodiversity inferences from eDNA.
ORIGINAL ARTICLES
DNA from dives: Species detection of humpback whales (Megaptera novaeangliae) from flukeprint eDNA
- First Published: 12 March 2024
Comparing cost, effort, and performance of environmental DNA sampling and trapping for detecting an elusive freshwater turtle
- First Published: 12 March 2024
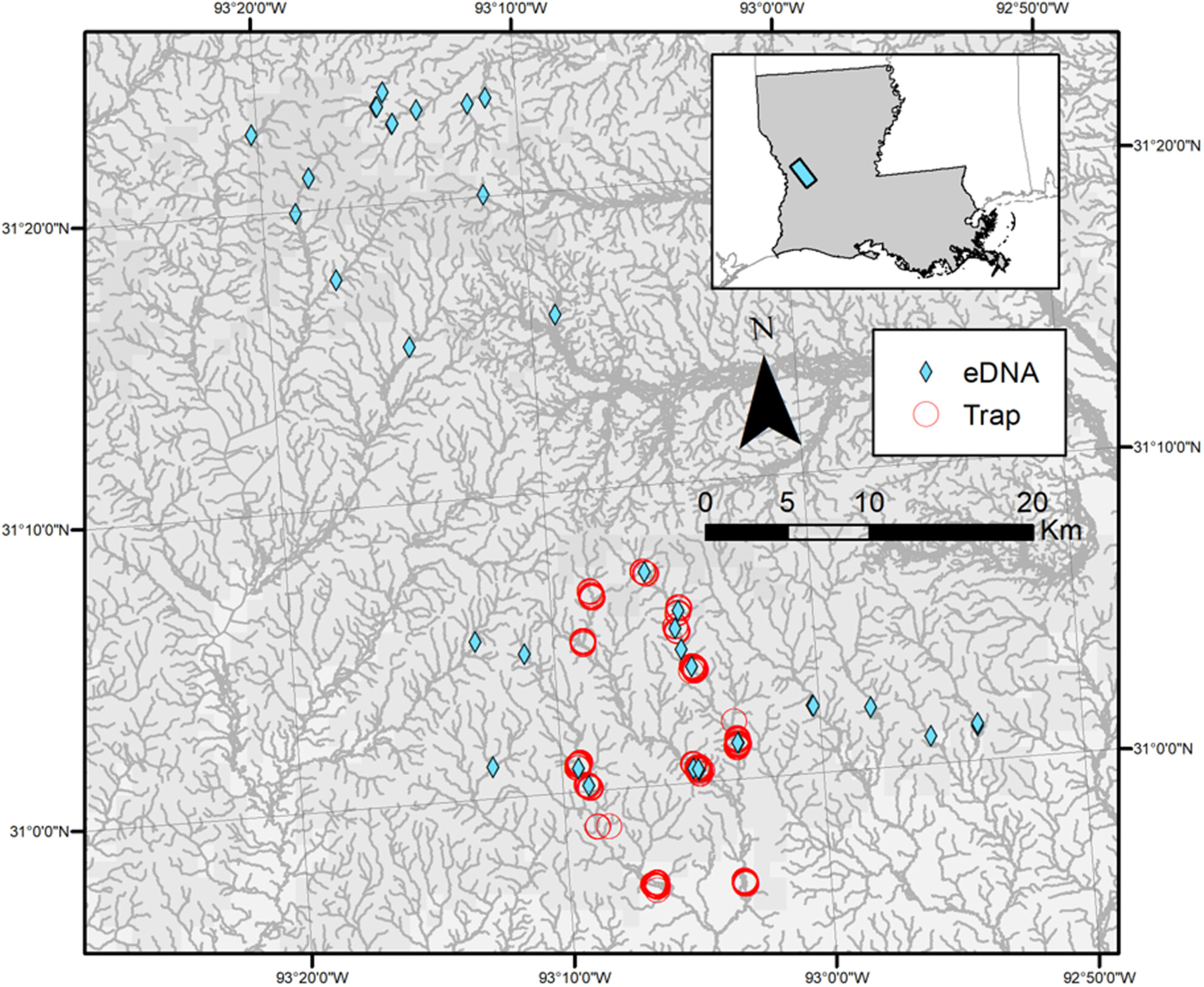
Successful integration of a new method into a standard management regime requires understanding both its relative efficacy and cost-effectiveness when compared against conventional sampling methods. We successfully detected M. temminckii in Louisiana streams using both eDNA surveys and conventional trapping with eDNA surveys yielding 5.55 times higher detection rates, 40% fewer labor hours per detection, and 84% more streams surveyed per year than conventional trapping. Our results suggest that eDNA presents a promising and cost-efficient technique for M. temminckii surveys.
METHODS
Environmental DNA metabarcoding of pan trap water to monitor arthropod-plant interactions
- First Published: 12 March 2024

The plant resources supporting arthropods in natural and agricultural landscapes are of increasing interest, largely because of the global decline in arthropod abundance and species diversity, as well as the increasing recognition of wild native pollinators as important contributors to crop yields. We demonstrate that eDNA metabarcoding of pan trap water can be used to identify the plant resources arthropods rely on. This approach complements long-standing survey methods and may potentially improve the characterization of plant–animal interactions in agroecosystems.
ORIGINAL ARTICLES
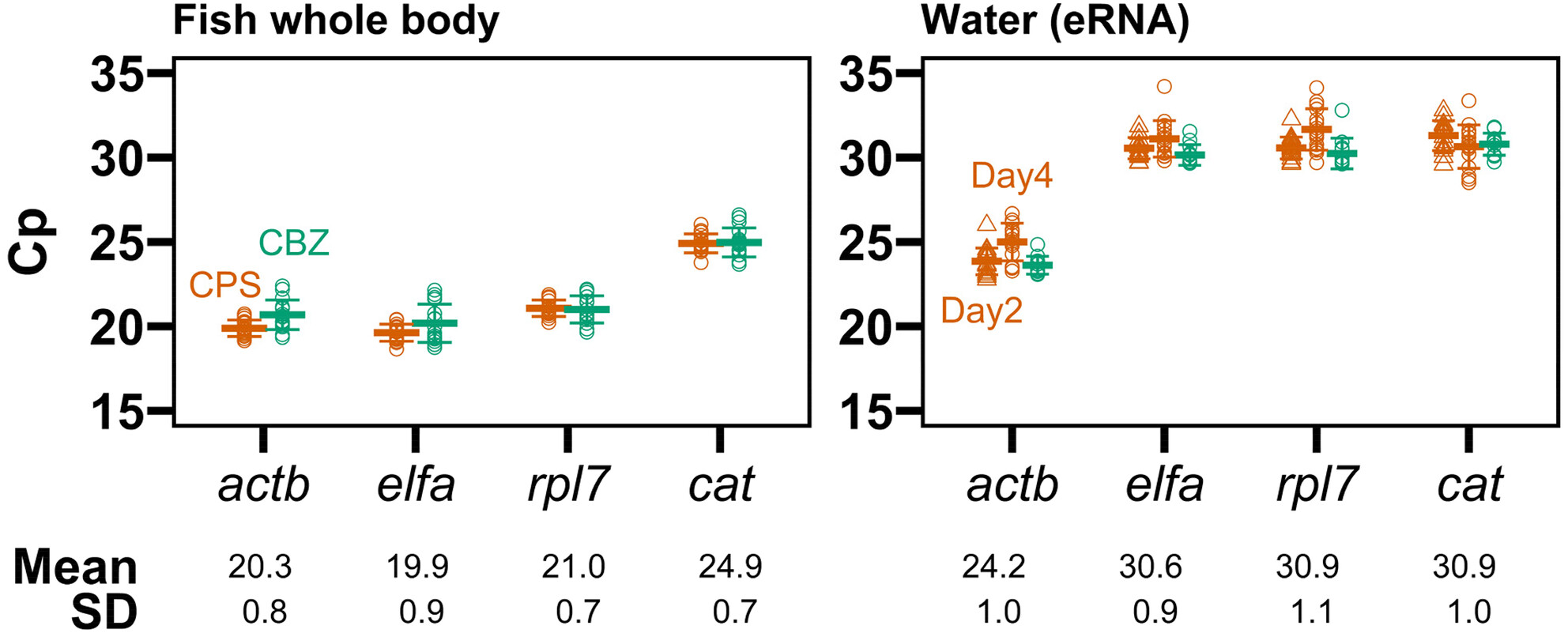
Relative gene expression analysis of catalase in environmental RNA from Japanese medaka exposed to toxic chemicals
- First Published: 12 March 2024
Tide impacts the dispersion of eDNA from nearshore net pens in a dynamic high-latitude marine environment
- First Published: 12 March 2024

In this study, we make use of hatchery net pens containing >46 million juvenile chum salmon (Oncorhynchus keta) in nearshore Southeast Alaska to test for dispersion of eDNA and the effects of tide. We collected and filtered surface water every 80 m along a 2 km transect to test eDNA attenuation over surface distance during incoming and outgoing tides on a single day. We found that surface samples showed a consistent signal of decreasing chum salmon eDNA across the 2 km transect, with the majority of eDNA detections within 1.5 km of the net pens and higher concentrations of DNA throughout during incoming tide.
Oomycete communities are influenced by land use and disease status in Christmas tree production in Southern Québec, Canada
- First Published: 15 March 2024
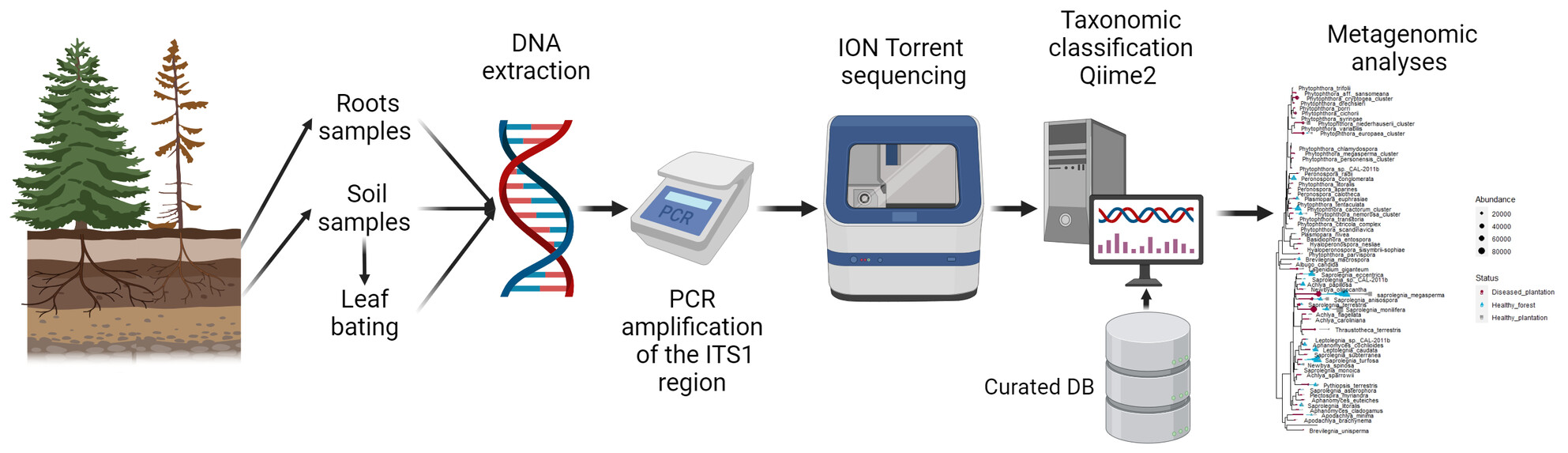
We use a metabarcoding approach to describe oomycetes' diversity and report that anthropogenic activities shape their diversity, while plantations can act as a gateway for invading natural forests. Our results suggested that the P. europaea cluster might already have crossed this boundary, advocating the importance of improved surveillance of oomycete diversity in various environments.
Proteinase K is not essential for marine eDNA metabarcoding
- First Published: 26 March 2024
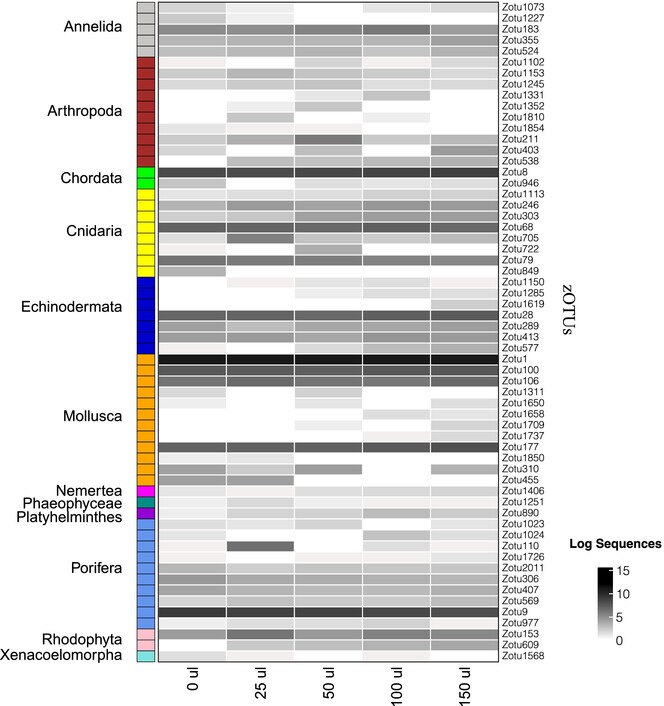
The manuscript examines whether Proteinase K (ProK), a standard and expensive ingredient in DNA extraction protocols, is necessary for protein-limited marine eDNA samples. We tested the effect of varying levels of ProK on DNA yield, ZOTU diversity, community composition and found no significant differences in diversity metrics among ProK quantities. Our results suggest that rare ZOTUs and eDNA patchiness are driving overall community differences as opposed to extraction ingredients.
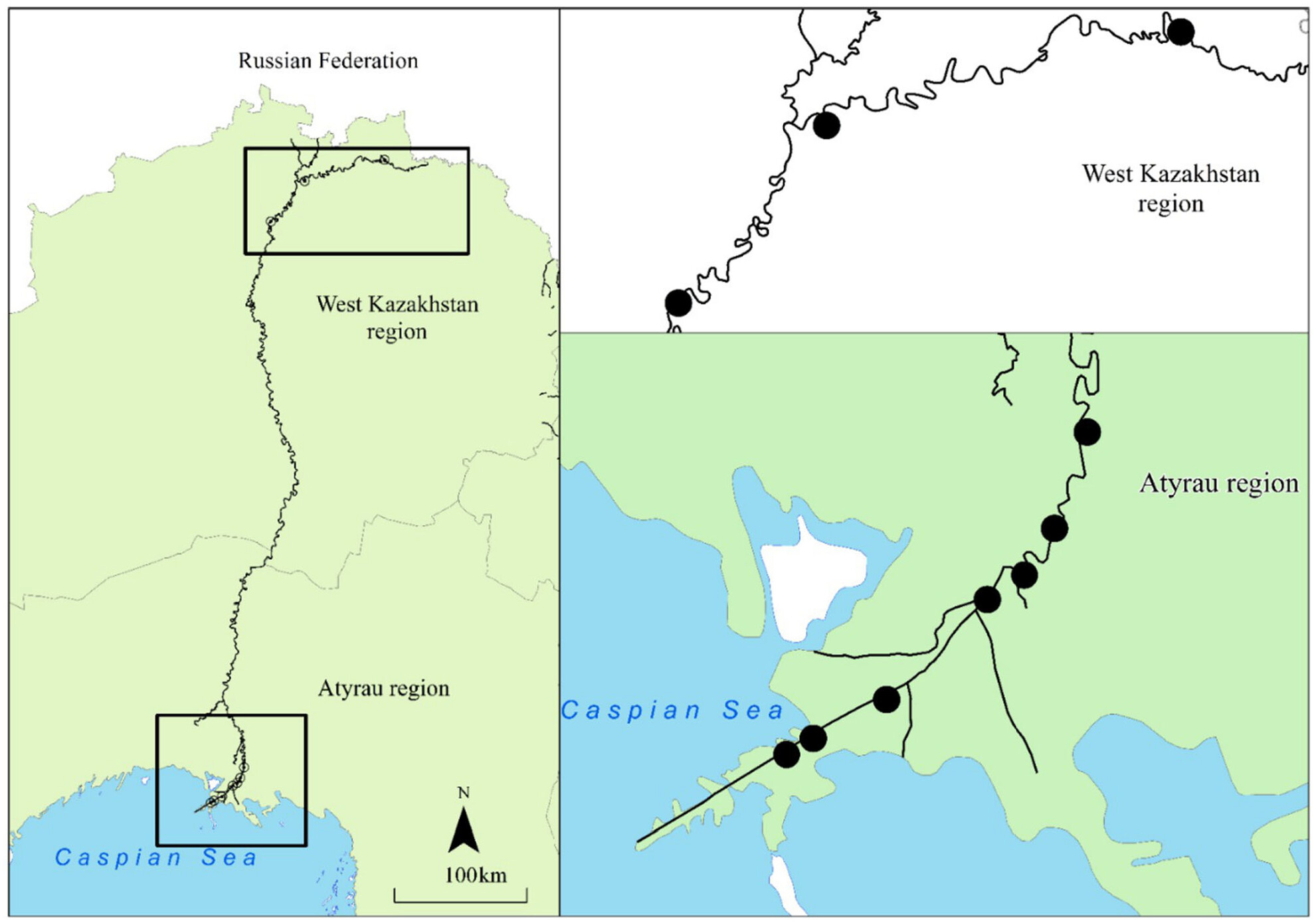
Environmental DNA reveals the ecology and seasonal migration of a rare sturgeon species in the Ural River
- First Published: 28 March 2024
SPECIAL ISSUE ARTICLES
Lessons learned from eDNA adoption in the management of bigheaded carps in Chicago IL USA Area Waterways
- First Published: 28 March 2024
Lessons form the early application of eDNA to detect invasive bigheaded carps in the Chicago Area Waterway System are relevant to accelerating the application of eDNA to other policy needs. Collaborative governance models, including the private sector, are likely to increase the societal benefits of eDNA for natural resource management.
ORIGINAL ARTICLES
Environmental DNA–RNA dynamics provide insights for effective monitoring of marine invasive species
- First Published: 28 March 2024
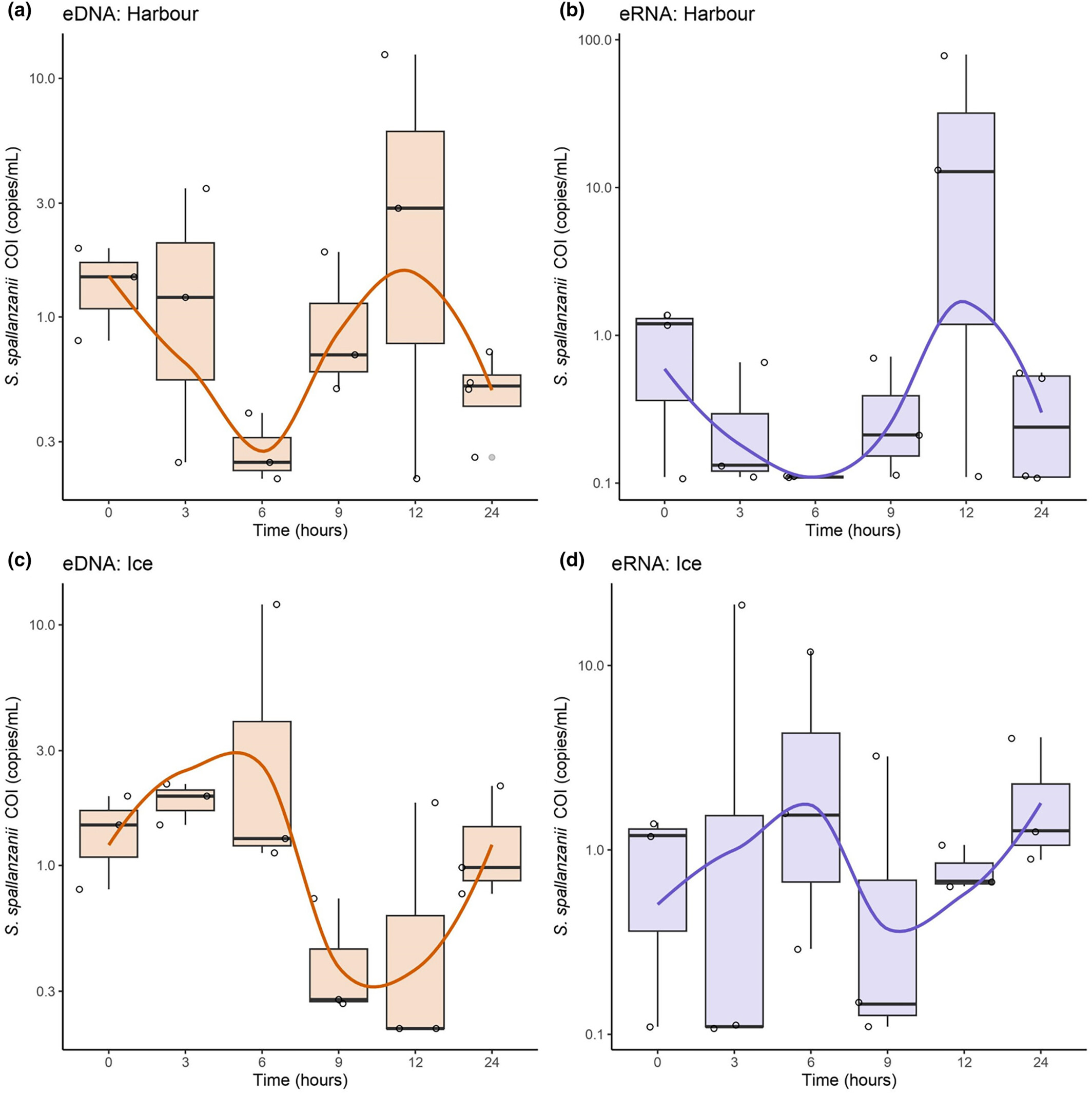
Environmental DNA and RNA can serve as molecular tools for biodiversity monitoring, yet questions about their persistence and dynamics within the field and in storage conditions remain. In our study seawater collected near a Sabella spallanzanii colony, was deployed in dialysis bags in a marina, with a subset of bags being stored on ice. Species-specific ddPCR results defied traditional decay models, revealing inconsistent eDNA/eRNA signal detection patterns over 24 h. Bioanalyzer results revealed an increase in larger fragments in field conditions, and preliminary results using long-read sequencing by Oxford Nanopore Technologies uncovered microbial eDNA growth in in-field samples, while ice-stored samples showed limited growth. These findings provide insights into total eDNA dynamics and its potential impact on targeted eDNA concentration and detection.
CORRECTION
Correction to “eDNA metabarcoding reveals riverine fish community structure and climate associations in northeastern Canada”
- First Published: 04 April 2024
ORIGINAL ARTICLES
Environmental DNA metabarcoding differentiates between micro-habitats within the rocky intertidal
- First Published: 06 April 2024
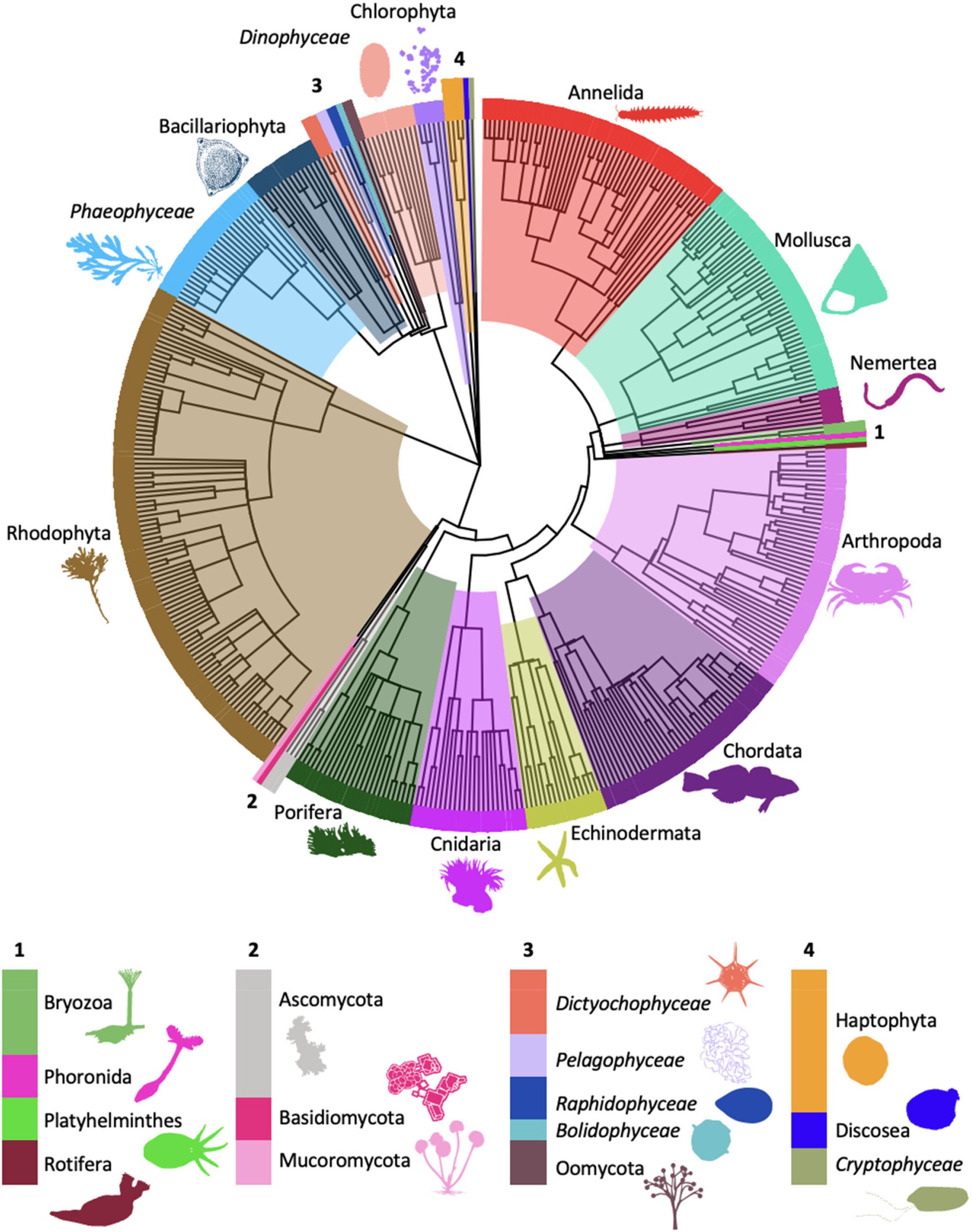
In this study, we collected eDNA samples from distinct micro-habitats in a rocky intertidal ecosystem over their exposure period in a tidal cycle. Using a well-established eukaryotic (cytochrome oxidase subunit I) metabarcoding assay, we detected 415 species across 28 phyla and identified unique eDNA signals from the different micro-habitats sampled. Our results demonstrate that eDNA biomonitoring can differentiate micro-habitats in the rocky intertidal only 40 m apart, that these differences reflect known ecology in the area, and that physical connectivity informs the degree of differentiation possible.
Design and partial validation of novel eDNA qPCR assays for three common North American tick (Arachnida: Ixodida) species
- First Published: 06 April 2024
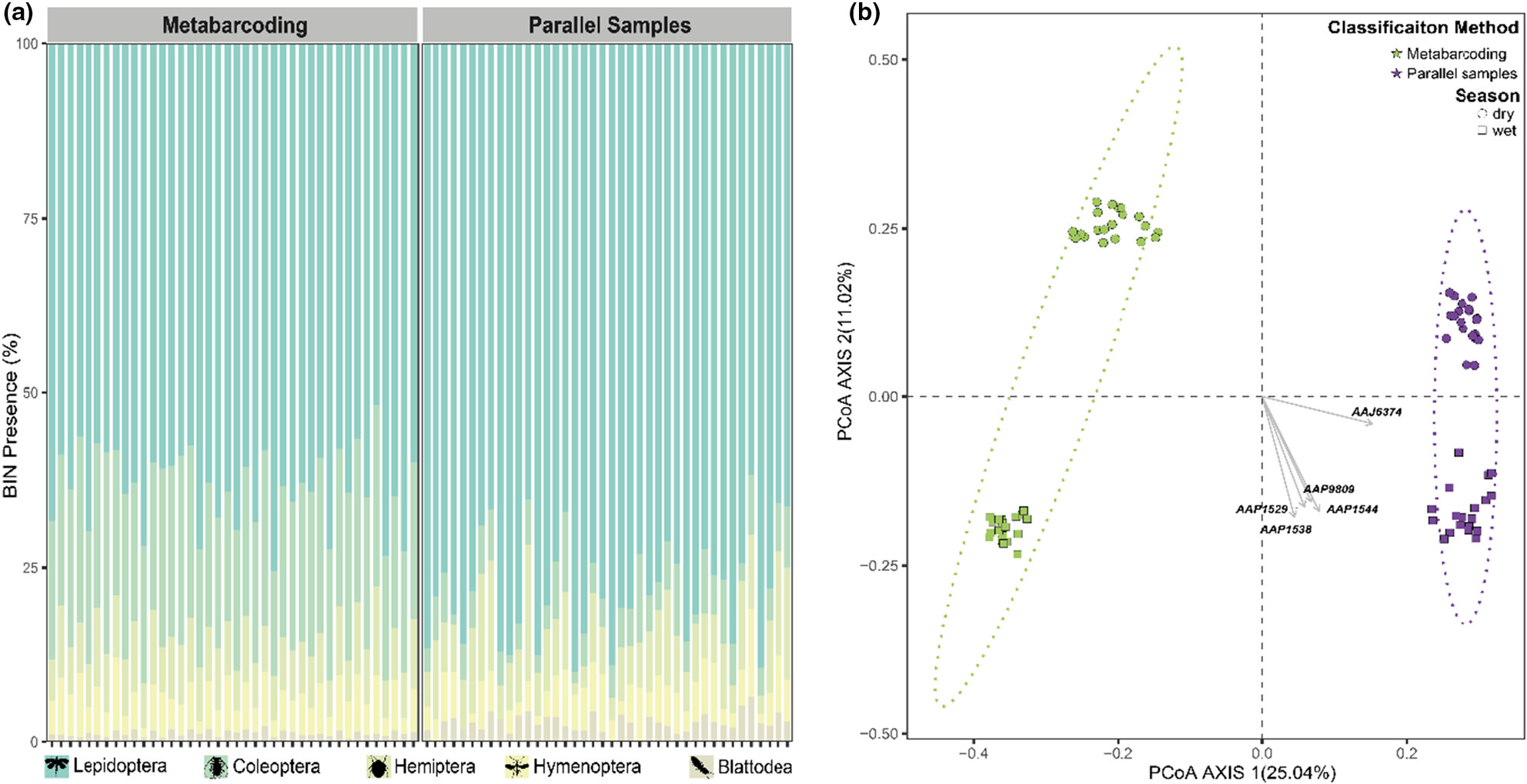
Illuminating arthropod diversity in a tropical forest: Assessing biodiversity by automatic light trapping and DNA metabarcoding
- First Published: 06 April 2024
Quantifying the effect of water quality on eDNA degradation using microcosm and bioassay experiments
- First Published: 15 April 2024
CORRECTION
Correction to “Single metabarcoding multiplex captures community-level freshwater biodiversity and beyond”
- First Published: 15 April 2024
METHODS
Recovering short DNA fragments from minerals and marine sediments: A comparative study evaluating lysis and isolation approaches
- First Published: 15 April 2024
Streamside detection of Chinook salmon (Oncorhynchus tshawytscha) environmental DNA with CRISPR technology
- First Published: 15 April 2024
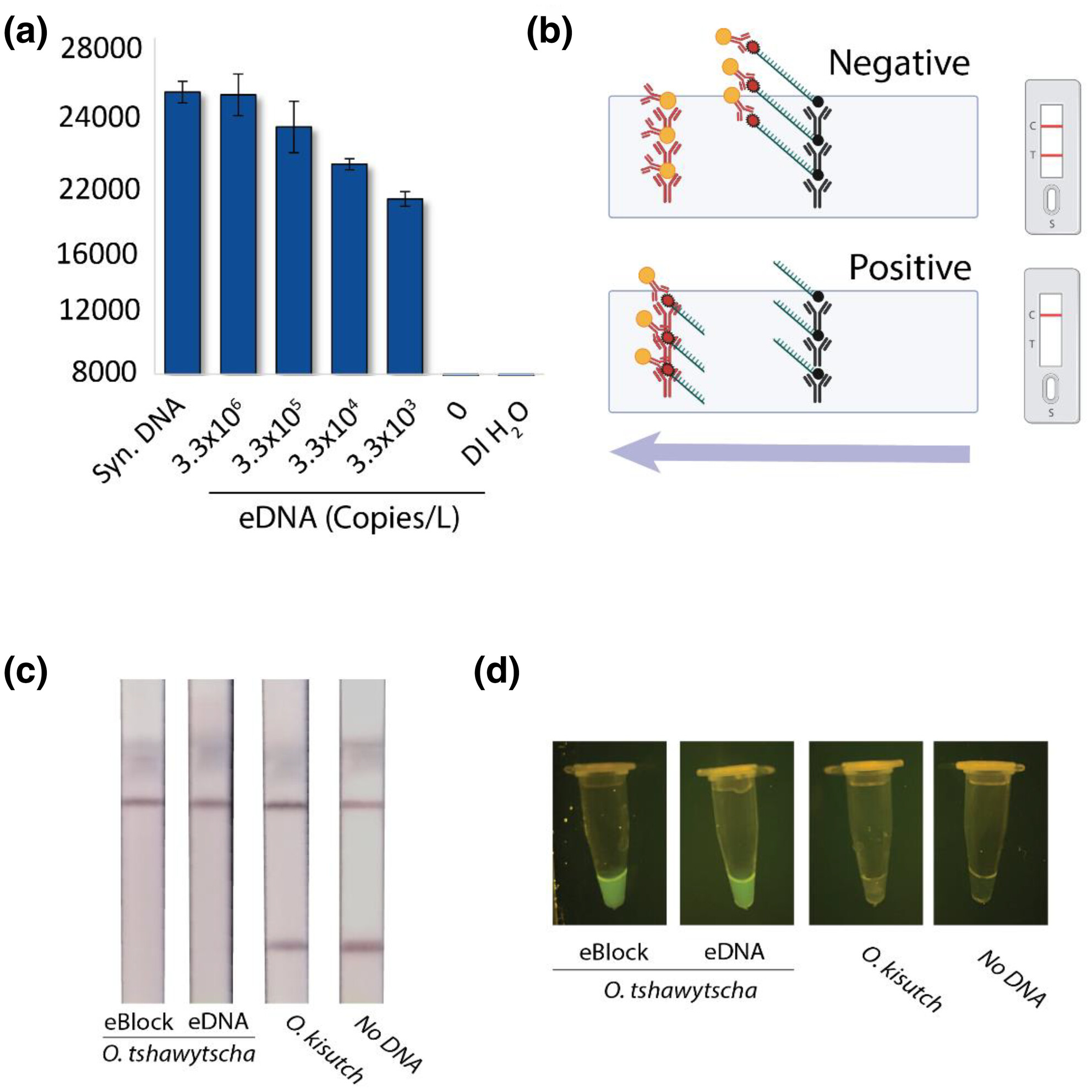
Environmental DNA (eDNA) is a powerful tool for resource management and conservation but currently requires specialized expertise and laboratory conditions to perform. The research seeks to expand the use of eDNA to field biologists by developing a streamside CRISPR/Cas12a detection technology for Chinook salmon eDNA. The method proved to be sensitive and specific and was able to successfully detect Chinook salmon in stream samples from the Snake River in the USA.
ORIGINAL ARTICLES
Dietary partitioning among three cryptobenthic reef fish mesopredators revealed by visual analysis, metabarcoding of gut content, and stable isotope analysis
- First Published: 24 April 2024
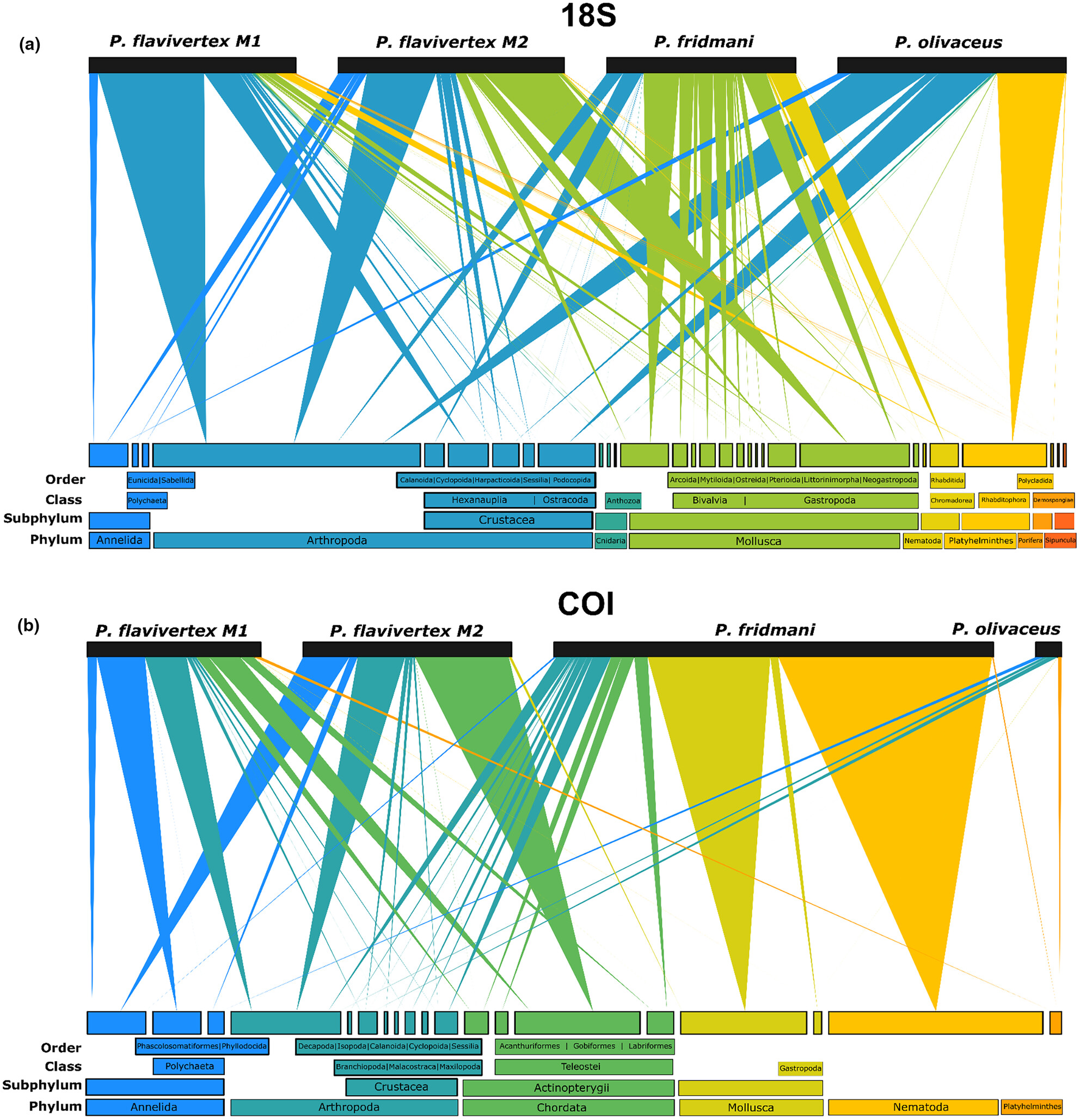
This study provides the first evidence of dietary partitioning of three common pseudochromid species in the Red Sea using a combination of (i) visual stomach content analysis, (ii) stomach content DNA metabarcoding (18S, COI), and (iii) stable isotope analysis (δ15N, δ13C). Our results revealed that Pseudochromis flavivertex, P. fridmani, and P. olivaceus present different dietary specializations and low dietary niche overlap among species. Our analysis highlights the importance of combining several approaches (short-term: visual analysis and DNA metabarcoding; and long-term: isotope analysis) when assessing the feeding habits of coral reef fishes, as they provide complementary information necessary to delimit their niches and understand the role that small mesopredators play in coral reef ecosystems.
Large-scale validation of 46 invasive species assays using an enhanced in silico framework
- First Published: 24 April 2024
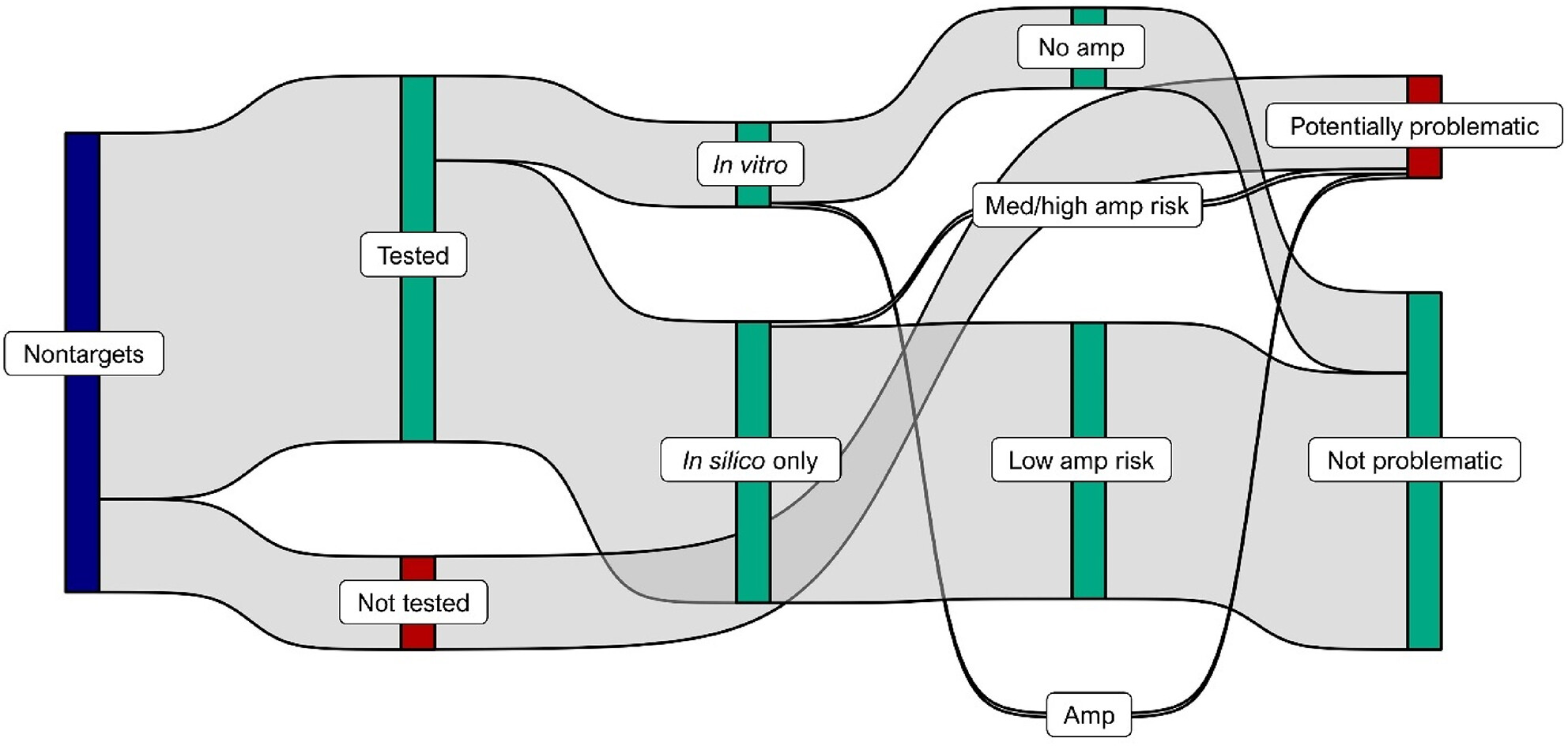
It can be challenging to ensure that eDNA assays are specific to their intended targets, particularly when biodiversity is high and the geographic area is large. Here, we use an accurate machine-learning-based PCR model to streamline validation of 46 qPCR assays targeting invasive species throughout the continental United States. We present an overview of our validation framework and trends in assay performance, along with detailed records of validation for each assay in an appendix.
METHODS
Generic qPCR assays for quality control in environmental DNA research
- First Published: 24 April 2024
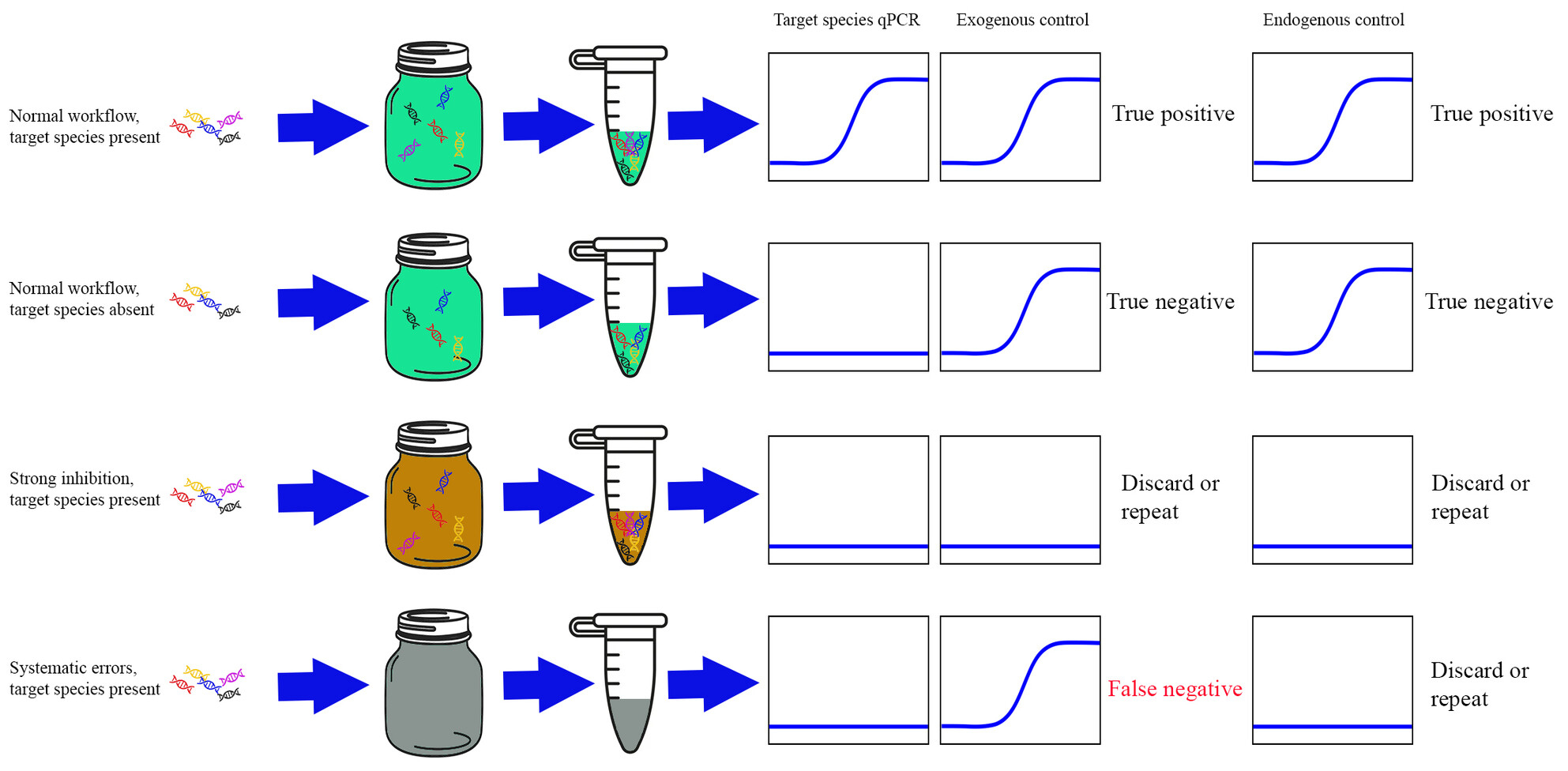
We developed two independent sets of generic quality control qPCR assays (QCqPCR) targeting abundant endogenous plant DNA in the environment. Unlike exogenous controls, our QCqPCR assays enable the monitoring of the entire eDNA workflow. We recommend incorporating either one of the QCqPCR assays into qPCR-based eDNA analysis to detect potential false negatives and improve the reliability of eDNA surveys.
ORIGINAL ARTICLES
The role of cultivated versus wild seeds in the diet of European turtle doves (Streptopelia turtur) across European breeding and African wintering grounds
- First Published: 24 April 2024
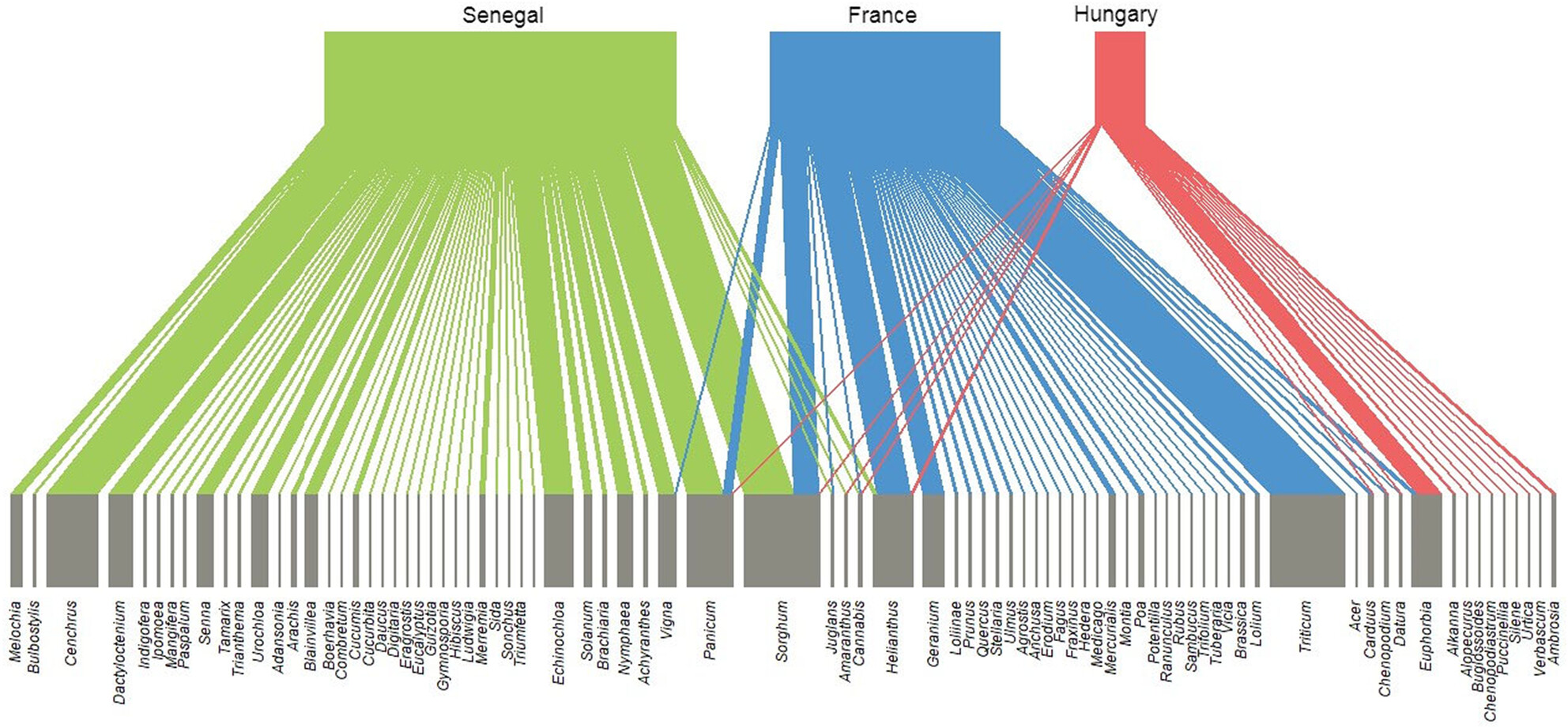
Using metabarcoding, we analyze the diet of the European turtle dove (Streptopelia turtur) in different sites, both breeding and wintering. We identified a diverse range of taxa in the diet and little overlap in the food consumed in breeding and wintering grounds. The roles of wild and cultivated food in the diet and implications for species management were considered.
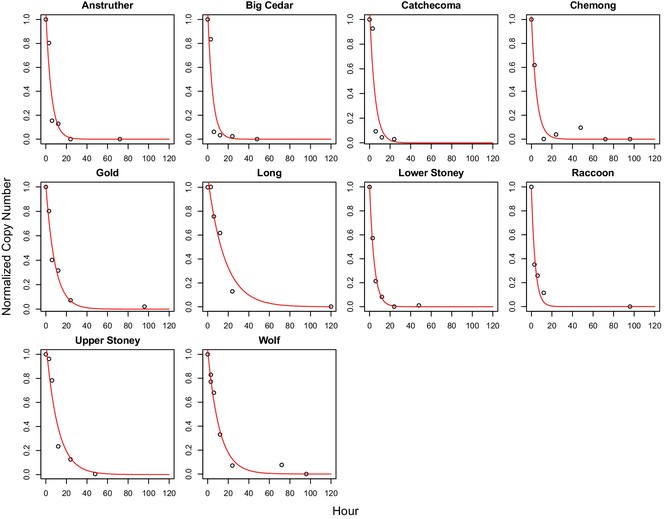
Environmental DNA dynamics of three species of unionid freshwater mussels
- First Published: 24 April 2024

We measured eDNA shedding and degradation rates of three freshwater mussel species, including one federally endangered species. We tested how biomass, feeding, and temperature affected shedding rate and found shedding rates were generally low for mussels compared with previous reports of shedding rates in fish. Degradation rates of eDNA were similar to those reported in the literature for other taxa.