Journal list menu
Export Citations
Download PDFs
ISSUE INFORMATION
EDITORIALS
Improving “may contain” labels: A call to team up and share data
- Pages: 1409-1412
- First Published: 12 January 2024
Nature's pharmacy at risk: Unveiling the hidden molecular secrets of phytotherapy in the age of globalization
- Pages: 1413-1415
- First Published: 03 February 2024
Is there a direct link between skin barrier dysfunction and asthma?
- Pages: 1416-1418
- First Published: 13 February 2024
EAACI POSITION PAPER
AllergoOncology: Biomarkers and refined classification for research in the allergy and glioma nexus—A joint EAACI-EANO position paper
- Pages: 1419-1439
- First Published: 24 January 2024
REVIEW ARTICLES
Tick bites, IgE to galactose-alpha-1,3-galactose and urticarial or anaphylactic reactions to mammalian meat: The alpha-gal syndrome
- Pages: 1440-1454
- First Published: 09 January 2024
Impact of climate change on atopic dermatitis: A review by the International Eczema Council
- Pages: 1455-1469
- First Published: 24 January 2024
The skin microbiome in pediatric atopic dermatitis and food allergy
- Pages: 1470-1484
- First Published: 03 February 2024
Skin, gut, and lung barrier: Physiological interface and target of intervention for preventing and treating allergic diseases
- Pages: 1485-1500
- First Published: 04 March 2024
Treatment of atopic dermatitis: Recently approved drugs and advanced clinical development programs
- Pages: 1501-1515
- First Published: 08 January 2024
ORIGINAL ARTICLES
Asthma and Lower Airway Disease
Transcriptomic evaluation of skin tape-strips in children with allergic asthma uncovers epidermal barrier dysfunction and asthma-associated biomarkers abnormalities
- Pages: 1516-1530
- First Published: 20 February 2024
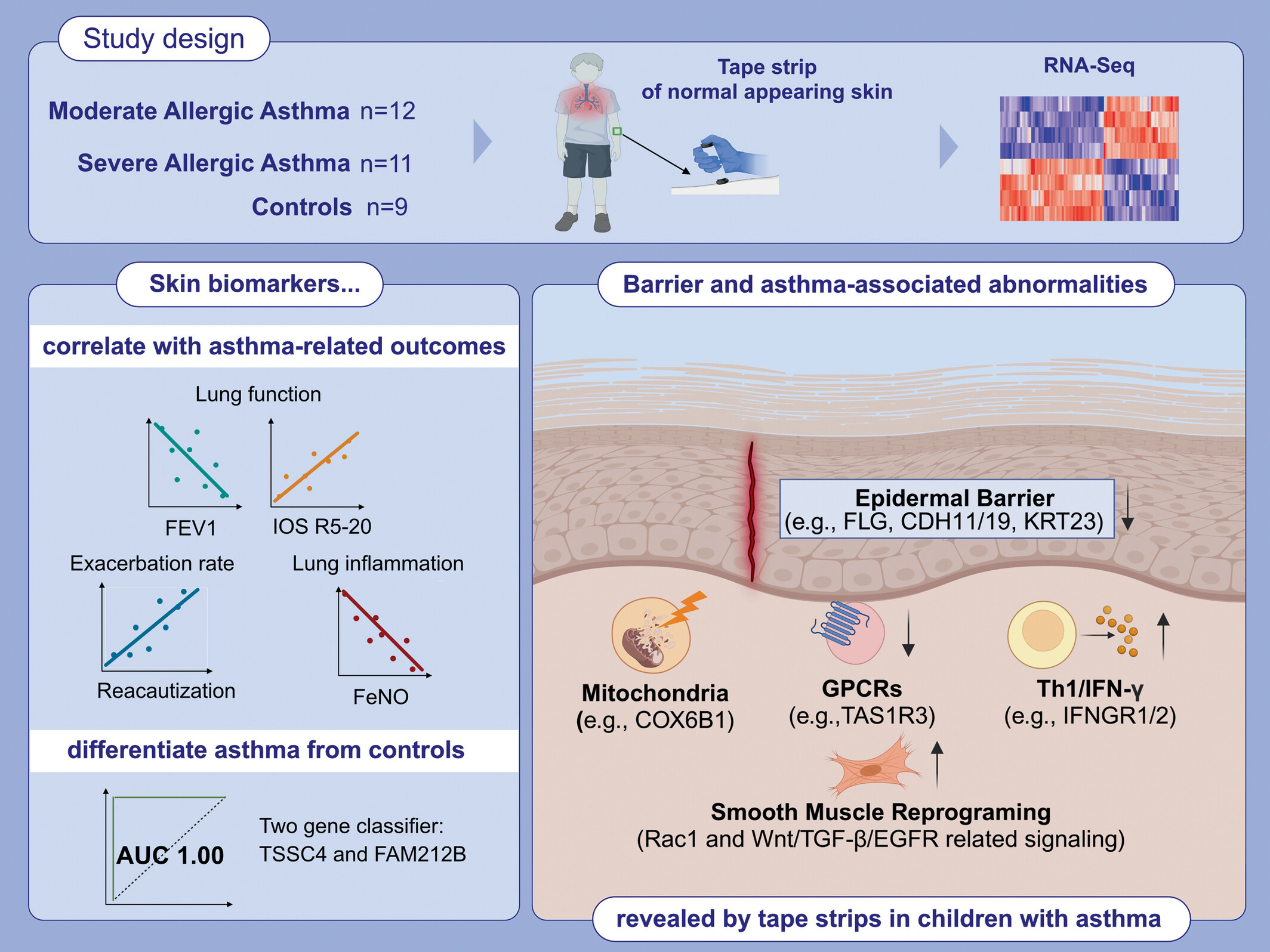
Using RNA-seq profiling, we characterized the normal-appearing skin of children with moderate allergic asthma and severe allergic asthma in comparison to healthy controls. Tape-strips of normal appearing skin from children with moderate allergic asthma and severe allergic asthma depict dysregulation of genes associated with epithelial barrier, lung extracellular milieu and correlate with asthma severity. A two-gene classifier, TSSC4 and FAM212B, differentiate asthmatic children from HCs with 100% accuracy.Abbreviations: AUC, area under the curve; CDH, cadherin; COX6B1, cytochrome c oxidase 6B1; EGFR, epidermal growth factor receptor; FAM212B, family with sequence similarity 212, member B; FeNO, fractional exhaled nitric oxide; FEV1; forced expiratory volume in 1 second; FLG, filaggrin; GPCRs, G protein-coupled receptors; IFN, interferon; IOS, impulse oscillometry; IOS-R5-20, a measure of peripheral airway resistance; KRT, keratin; RNA-seq, RNA sequencing; TAS1R3; taste 1 receptor member 3; TGF, transforming growth factor; Th, T helper; TSSC4, tumor suppressing subtransferable candidate 4.
Atopic Dermatitis, Urticaria and Skin Disease
Epidermal barrier impairment predisposes for excessive growth of the allergy-associated yeast Malassezia on murine skin
- Pages: 1531-1547
- First Published: 22 February 2024
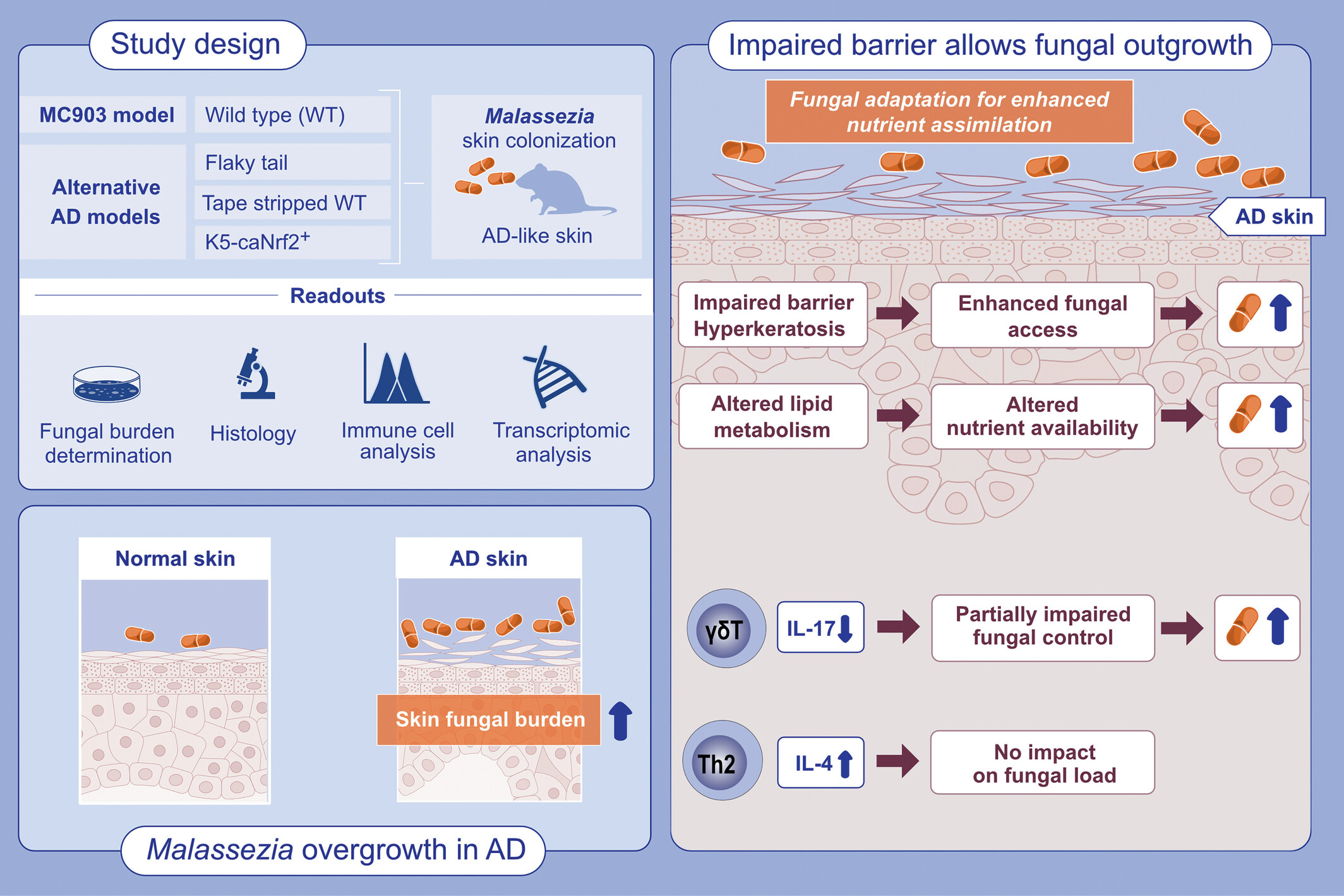
The abundant commensal yeast Malassezia overgrows in AD-like skin. Using four different experimental models of AD, we show that Malassezia acquires enhanced fitness in the barrier-impaired and metabolically dysregulated skin. The lipid-dependent yeast adapts to the AD-like skin for enhanced nutrient assimilation.Abbreviations: AD, atopic dermatitis; WT, wildtype.
RNA-sequencing of paired tape-strips and skin biopsies in atopic dermatitis reveals key differences
- Pages: 1548-1559
- First Published: 13 March 2024

Tape strips yielded a lower yield of RNA and fewer sequences mapping to known genes than biopsies. Paired sampling from the same skin (AD or no AD) identified differential detection of thousands of genes between tape strips and biopsies. Tape strips performed better than biopsies for identifying differentially expressed genes in AD.
Targeting IL-13 with tralokinumab normalizes type 2 inflammation in atopic dermatitis both early and at 2 years
- Pages: 1560-1572
- First Published: 02 April 2024
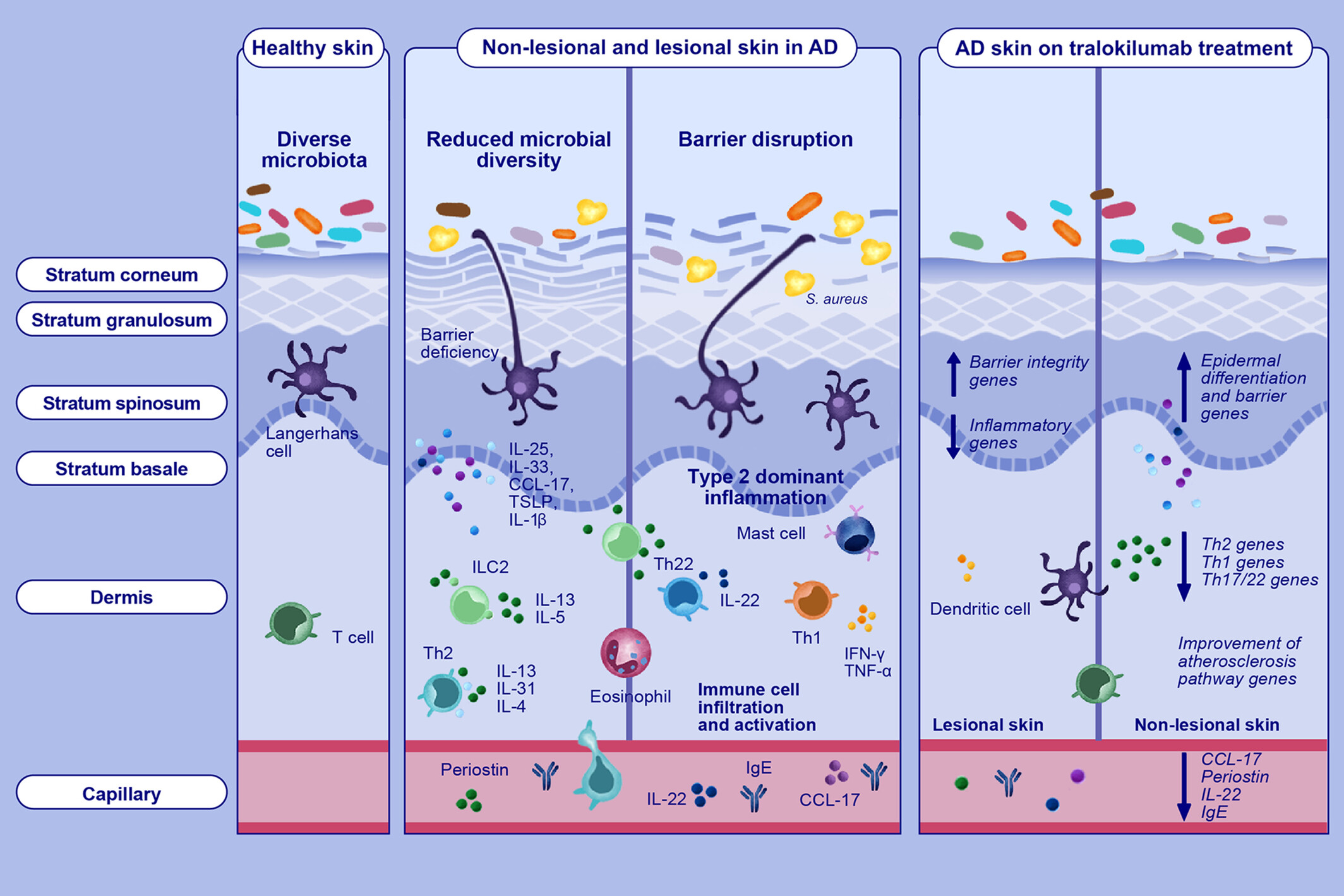
This study evaluated early and 2-year impacts of IL-13 neutralization on skin and serum biomarkers following tralokinumab treatment in adults with moderate-to- severe AD. Specific targeting of IL-13 with tralokinumab improved epidermal pathology and normalized expression of key biomarkers in serum and lesional skin in patients with moderate-to-severe AD. These data further support the central role of IL-13 in driving AD pathology and highlight that inhibition of IL-13 alone is sufficient for normalizing the molecular phenotype of AD. Abbreviations: AD, atopic dermatitis; CCL-17, CC-motif chemokine ligand 17; IFN-γ, interferon γ; IgE, immunoglobulin E; IL, interleukin; ILC2, group 2 innate lymphoid cells; Th, T helper cell; TNF-α; tumor necrosis factor α; TSLP, thymic stromal lymphopoietin.
Inhibition of RNase 7 by RNase inhibitor promotes inflammation and Staphylococcus aureus growth: Implications for atopic dermatitis
- Pages: 1573-1583
- First Published: 19 April 2024
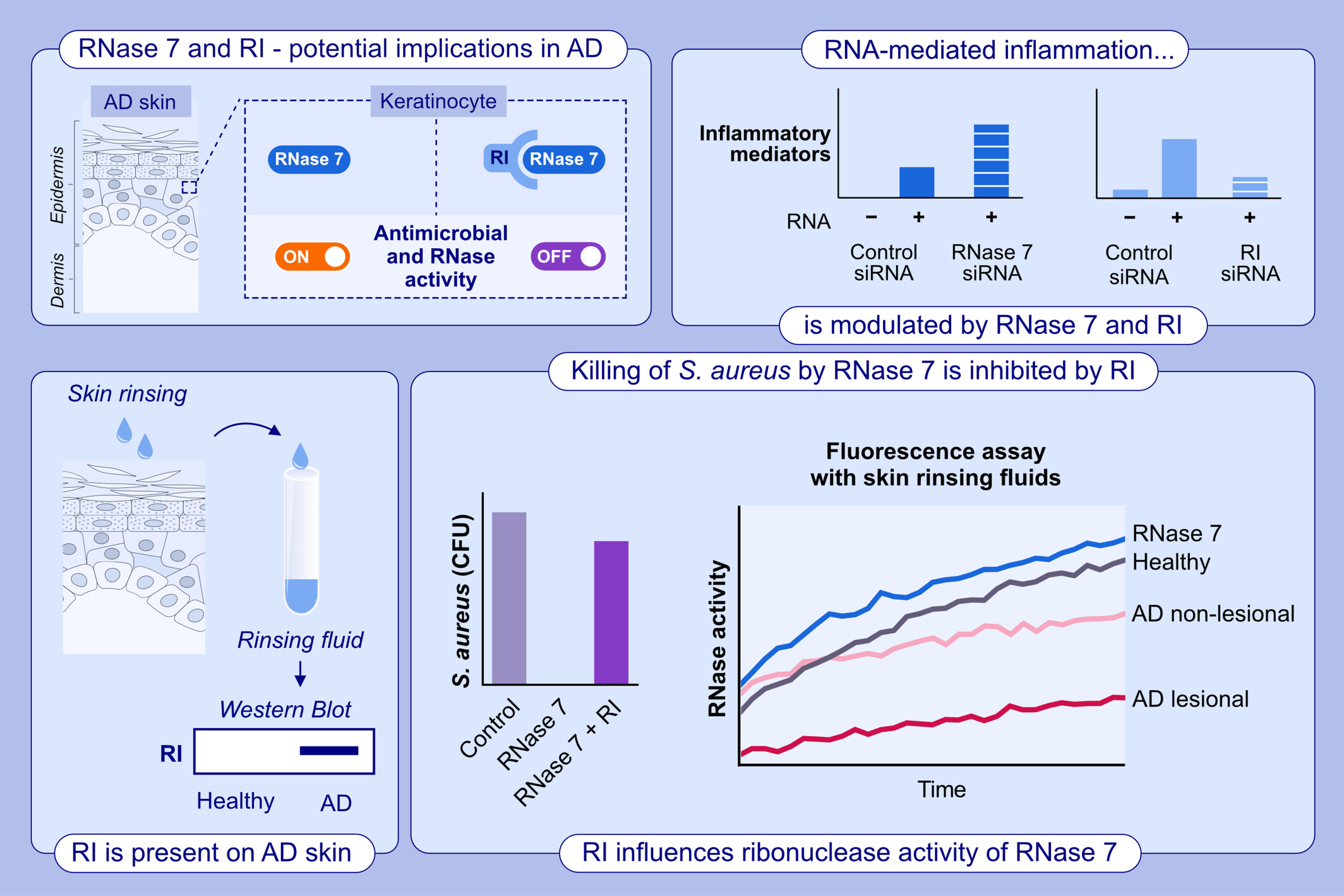
The lesional skin of AD patients releases increased amounts of RNase inhibitor (RI). Host RI binds to RNase7 and blocks its ribonuclease and antimicrobial activity. This inactivation has an enhancing effect on the RNA-mediated pro-inflammatory response and Staphylococcus aureus growth. These new data reveal a previously unknown role of the RNase7–RI interaction in AD.Abbreviations: AD, atopic dermatitis; CFU, colony forming units; RNA, ribonucleic acid; RI, RNase inhibitor; S. aureus, Staphylococcus aureus; siRNA, small interfering RNA.
Mapping the immune cell landscape of severe atopic dermatitis by single-cell RNA-seq
- Pages: 1584-1597
- First Published: 31 May 2024
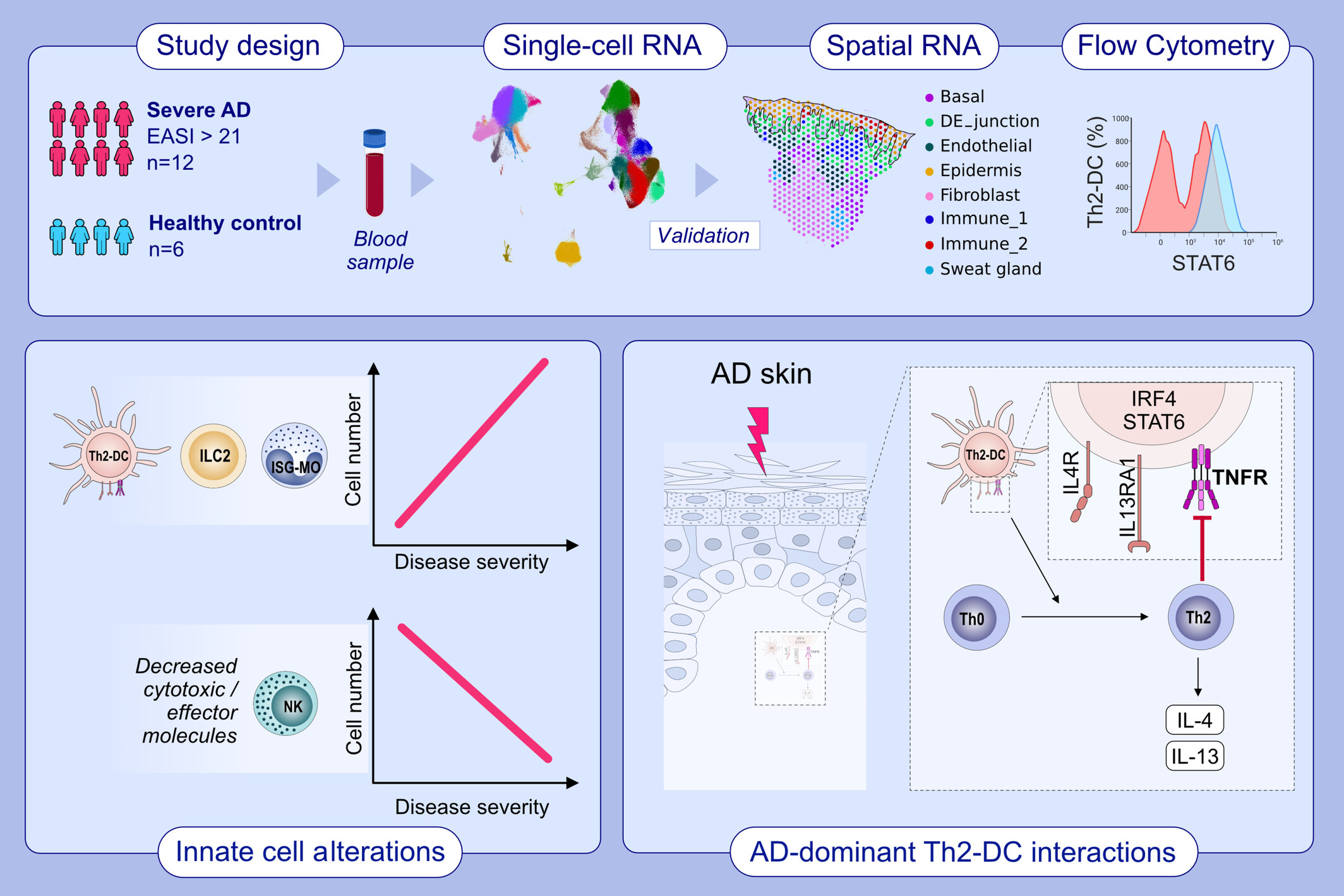
We analyzed blood samples from 12 patients with severe atopic dermatitis (EASI > 21) using single-cell RNA sequencing. Additionally, we validated our findings with skin single-cell spatial sequencing and flow cytometry. Our findings revealed a strong correlation between disease severity and the expansion of ILC2s and ISG-monocytes. Conversely, the number of natural killer (NK) cells was decreased in the patients, and the expression of cytotoxic/effector molecules in NK cells was negatively correlated with disease severity. Moreover, our study identified severe AD-dominant Th2-DCs that exhibited features of Th2-priming DC signals and interacted with Th2 cells residing in AD lesional skin.Abbreviations: AD, atopic dermatitis; DC, dendritic cell; DE_junction, dermis epidermis junction; EASI, eczema area and severity index; IL, interleukin; ILC2, group 2 innate lymphoid cell; IRF4, interferon regulatory factor 4; ISG, interferon-stimulated genes; MO, monocyte; NK, natural killer cell; STAT6, signal transducer and activator of transcription 6; TNFR, tumor necrosis factor receptor.
LETTERS
Epidemiology of severe childhood asthma in Japan: A nationwide descriptive study
- Pages: 1598-1602
- First Published: 07 January 2024
Activation of basophils in children with food allergies by blood from donors ingesting the corresponding food
- Pages: 1602-1605
- First Published: 11 January 2024
Butyrate dietary supplementation promotes tolerant responses in a Pru p 3-anaphylactic mouse model
- Pages: 1605-1608
- First Published: 23 January 2024
CXCL17 induces activation of human mast cells via MRGPRX2
- Pages: 1609-1612
- First Published: 26 January 2024
Efficacy and safety of topical Streptococcus postbiotic emollient in adolescents and adults with mild-to-moderate atopic dermatitis: A randomized, double-blind, vehicle-controlled trial
- Pages: 1612-1616
- First Published: 04 March 2024
Meta-analysis in allergy—Statistical recommendations
- Pages: 1616-1618
- First Published: 29 February 2024
Neonatal skin dysbiosis to infantile atopic dermatitis: Mitigating effects of skin care
- Pages: 1618-1622
- First Published: 09 March 2024
Interleukin 31 transcription in a canine model of acute atopic dermatitis does not correlate with T-cell infiltration
- Pages: 1622-1625
- First Published: 26 March 2024
SM17, a new IL-17RB-targeting antibody, ameliorates disease progression in a mouse model of atopic dermatitis
- Pages: 1625-1628
- First Published: 09 April 2024
NEWS AND VIEWS
Legends of Allergy and Immunology
Legends of allergy and immunology: Hirohisa Saito
- Pages: 1629-1630
- First Published: 14 February 2024
Groundbreaking Discoveries in Allergy, Asthma and Immunology
Insights in intestinal immune tolerance: The role of the cleavage form of gasdermin D
- Pages: 1631-1632
- First Published: 25 February 2024
CORRESPONDENCES
Reply to “European academy of allergy and clinical immunology, food allergy, anaphylaxis guidelines group. EAACI guidelines: Anaphylaxis (2021 update)”
- Pages: 1633-1634
- First Published: 12 January 2024
Correspondence to “Short-course subcutaneous treatment with PQ Grass strongly improves symptom and medication scores in grass allergy1”
- Pages: 1635-1636
- First Published: 16 January 2024
Response to Correspondence to “Short-course subcutaneous treatment with PQ Grass strongly improves symptom and medication scores in grass allergy”
- Pages: 1637-1638
- First Published: 25 March 2024
CORRIGENDUM
Correction to “Alveolar macrophages are sentinels of murine pulmonary homeostasis following inhaled antigen challenge”
- Pages: 1639-1640
- First Published: 10 January 2024
ERRATUM
Correction to “Mind and skin: Exploring the links between inflammation, sleep disturbance and neurocognitive function in patients with atopic dermatitis”
- Page: 1641
- First Published: 25 February 2024






