Journal list menu
Export Citations
Download PDFs
ISSUE INFORMATION - COVER IMAGE
Cover Image, Volume 89, Issue 12
- Page: C1
- First Published: 24 November 2021
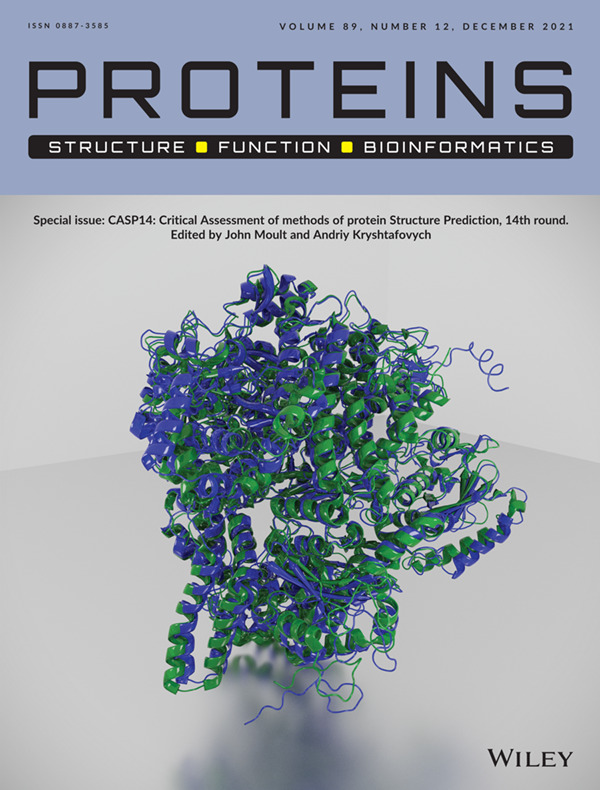
Front Cover: The cover image has been created by Clemens Meyer and Adam Cain from DeepMind. It shows the ground truth experimental structure for CASP14 target T1044 in green, together with the prediction of the AlphaFold2 team in blue.Target T1044 (PDB 6VR4) is the RNA polymerase of the crAss-like phage phi14:2, a large single-chain protein with 2180 residues. Its structure was published by Drobysheva et al. (Nature, 2020). Target T1044 was split into 9 domains for the CASP14 experiment (T1031, T1033, T1035, T1037, T1039, T1040, T1041, T1042, and T1043); a prediction for the full protein chain, including domain packing, is shown here.
TABLE OF CONTENT
Issue Information - Table of Content
- Pages: 1603-1606
- First Published: 24 November 2021
RESEARCH ARTICLES
Critical assessment of methods of protein structure prediction (CASP)—Round XIV
- Pages: 1607-1617
- First Published: 17 September 2021
Target classification in the 14th round of the critical assessment of protein structure prediction (CASP14)
- Pages: 1618-1632
- First Published: 04 August 2021
Computational models in the service of X-ray and cryo-electron microscopy structure determination
- Pages: 1633-1646
- First Published: 27 August 2021
Target highlights in CASP14: Analysis of models by structure providers
- Pages: 1647-1672
- First Published: 25 September 2021
Topology evaluation of models for difficult targets in the 14th round of the critical assessment of protein structure prediction (CASP14)
- Pages: 1673-1686
- First Published: 09 July 2021
High-accuracy protein structure prediction in CASP14
- Pages: 1687-1699
- First Published: 03 July 2021
Assessment of domain interactions in the fourteenth round of the Critical Assessment of Structure Prediction (CASP14)
- Pages: 1700-1710
- First Published: 29 August 2021
Applying and improving AlphaFold at CASP14
- Pages: 1711-1721
- First Published: 02 October 2021
Protein tertiary structure prediction and refinement using deep learning and Rosetta in CASP14
- Pages: 1722-1733
- First Published: 30 July 2021
Protein structure prediction using deep learning distance and hydrogen-bonding restraints in CASP14
- Pages: 1734-1751
- First Published: 30 July 2021
Assessing the utility of CASP14 models for molecular replacement
- Pages: 1752-1769
- First Published: 12 August 2021
REVIEW ARTICLE
Protein sequence-to-structure learning: Is this the end(-to-end revolution)?
- Pages: 1770-1786
- First Published: 13 September 2021
RESEARCH ARTICLES
Prediction of protein assemblies, the next frontier: The CASP14-CAPRI experiment
- Pages: 1800-1823
- First Published: 28 August 2021
Protein oligomer modeling guided by predicted interchain contacts in CASP14
- Pages: 1824-1833
- First Published: 29 July 2021
Modeling of protein complexes in CASP14 with emphasis on the interaction interface prediction
- Pages: 1834-1843
- First Published: 27 June 2021
Protein oligomer structure prediction using GALAXY in CASP14
- Pages: 1844-1851
- First Published: 06 August 2021
Evaluation of model refinement in CASP14
- Pages: 1852-1869
- First Published: 20 July 2021
Physics-based protein structure refinement in the era of artificial intelligence
- Pages: 1870-1887
- First Published: 22 June 2021
Assessing the accuracy of contact and distance predictions in CASP14
- Pages: 1888-1900
- First Published: 01 October 2021
When homologous sequences meet structural decoys: Accurate contact prediction by tFold in CASP14—(tFold for CASP14 contact prediction)
- Pages: 1901-1910
- First Published: 02 September 2021
Protein inter-residue contact and distance prediction by coupling complementary coevolution features with deep residual networks in CASP14
- Pages: 1911-1921
- First Published: 12 August 2021
Assessing the binding properties of CASP14 targets and models
- Pages: 1922-1939
- First Published: 09 August 2021
Assessment of protein model structure accuracy estimation in CASP14: Old and new challenges
- Pages: 1940-1948
- First Published: 29 July 2021
Assessment of prediction methods for protein structures determined by NMR in CASP14: Impact of AlphaFold2
- Pages: 1959-1976
- First Published: 24 September 2021
Continuous Automated Model EvaluatiOn (CAMEO)—Perspectives on the future of fully automated evaluation of structure prediction methods
- Pages: 1977-1986
- First Published: 12 August 2021
Modeling SARS-CoV-2 proteins in the CASP-commons experiment
- Pages: 1987-1996
- First Published: 30 August 2021
ISSUE INFORMATION - FORTHCOMING
ISSUE INFORMATION - COVER IMAGE
Cover Image, Volume 89, Issue 12
- Page: C4
- First Published: 24 November 2021

Reverse Cover: The cover image is based on the review Critical assessment of methods of protein structure prediction (CASP) – Round XIV by Andriy Kryshtafovych, Torsten Schwede, Maya Topf, Krzysztof Fidelis and John Moult, DOI 10.1002/prot.26237. The highlight of the CASP14 experiment was an outstanding performance of AlphaFold2 structure modeling method. The cover image shows the backbone agreement with experiment of the best models in CASP1 (1994), CASP5 (2002) and the latest three CASP experiments. The data are shown as linear trend lines; for the most recent round, individual target points are shown as circles. Performance in CASP14 (top black line) is very impressive, with accuracy approaching experimental accuracy for many targets (GDT_TS of 90 or more). This outstanding performance is dominated by AlphaFold2 models, although other groups also advanced substantially from CASP13 (dashed black line). The agreement metric, GDT_TS, is a multi-scale indicator of the closeness of the Cα atoms in a model to those in the corresponding experimental structure. Target difficulty is based on sequence and structure similarity to other proteins with known experimental structures.




