Journal list menu
Export Citations
Download PDFs
Front Cover, Volume 43, Issue 8
- Page: i
- First Published: 15 July 2022
Front Cover: The cover image is based on the Editorial Introduction Human mutation special issue on “variant effect prediction” by Ales Maver et al., https://doi.org/10.1002/humu.24429.
Human Mutation special issue on “Variant Effect Prediction"
- Pages: 973-975
- First Published: 15 July 2022
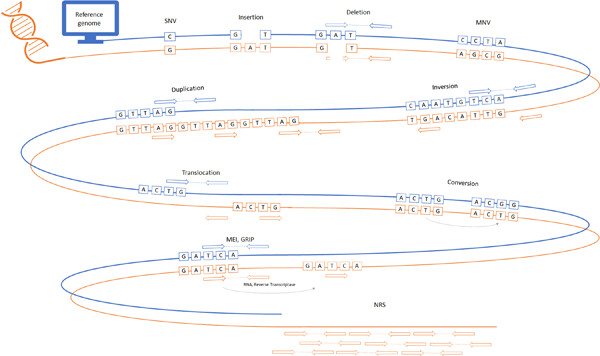
Variant calling: Considerations, practices, and developments
- Pages: 976-985
- First Published: 09 December 2021
Annotating and prioritizing genomic variants using the Ensembl Variant Effect Predictor—A tutorial
- Pages: 986-997
- First Published: 24 November 2021

The Ensembl Variant Effect Predictor (VEP) is a freely available, open-source tool for the annotation and prioritization of genomic variants. It maps variants to functionally relevant genomic regions and predicts their consequence. In this tutorial, we explain how to use the Ensembl VEP web interface and describe the available annotation options, including reference population frequency data, phenotype associations, variant citation data, and pathogenicity predictors.
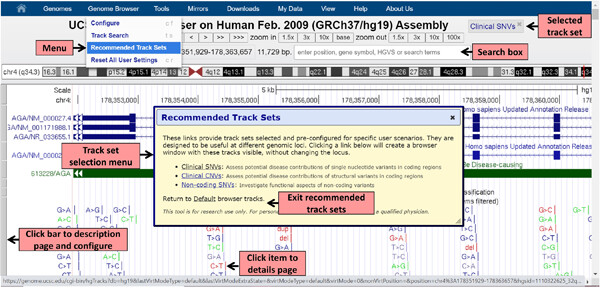
Variant interpretation: UCSC Genome Browser Recommended Track Sets
- Pages: 998-1011
- First Published: 28 January 2022
Graph
The new UCSC Recommended Track Sets are designed to facilitate the interpretation of variants in the genome, offering quick access to relevant and appropriately configured tracks, allowing visualization and easy comparison of data from multiple sources, and support researches in understanding the relationship between genetic variants and human disease in the course of testing patient specimens. “Supporting Information” is included.
Variant interpretation using population databases: Lessons from gnomAD
- Pages: 1012-1030
- First Published: 02 December 2021
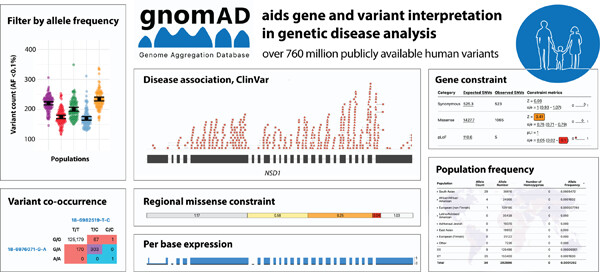
Reference population databases are critical in the interpretation of genomic variation for diagnosing rare disease, and supports the discovery of new disease–gene relationships. This review provides guidance for using the Genome Aggregation Database (gnomAD) browser and key features like allele frequency, per-base expression levels, constraint scores, and variant co-occurrence, for variant and gene interpretation in clinical and research analysis.
Utilizing ClinGen gene-disease validity and dosage sensitivity curations to inform variant classification
- Pages: 1031-1040
- First Published: 25 October 2021
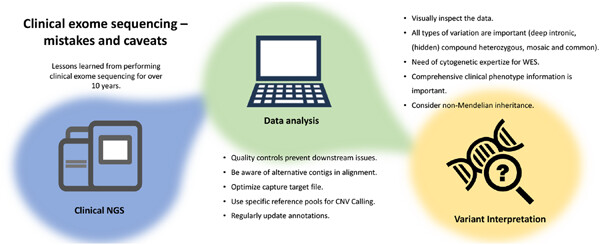
Clinical exome sequencing—Mistakes and caveats
- Pages: 1041-1055
- First Published: 22 February 2022
Guidelines for clinical interpretation of variant pathogenicity using RNA phenotypes
- Pages: 1056-1070
- First Published: 29 May 2022
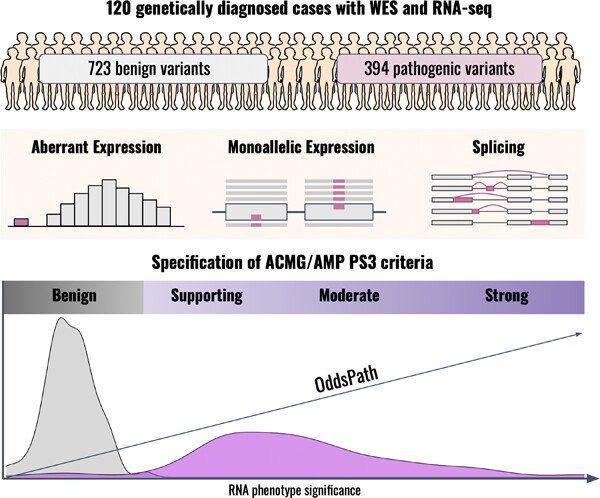
The purpose of these guidelines is to provide a formal integration of RNA phenotypes into the American College of Medical Genetics and Genomics and the Association for Molecular Pathology framework as functional evidence. These recommendations were also designed to serve as an educational resource for clinical laboratory geneticists to facilitate the clinical implementation of RNA-seq (RNA-sequencing).
Phenotype-driven approaches to enhance variant prioritization and diagnosis of rare disease
- Pages: 1071-1081
- First Published: 07 April 2022
Gene–disease relationship evidence: A clinical perspective focusing on ultra-rare diseases
- Pages: 1082-1088
- First Published: 09 March 2022
Adapting the ACMG/AMP variant classification framework: A perspective from the ClinGen Hemoglobinopathy Variant Curation Expert Panel
- Pages: 1089-1096
- First Published: 12 September 2021
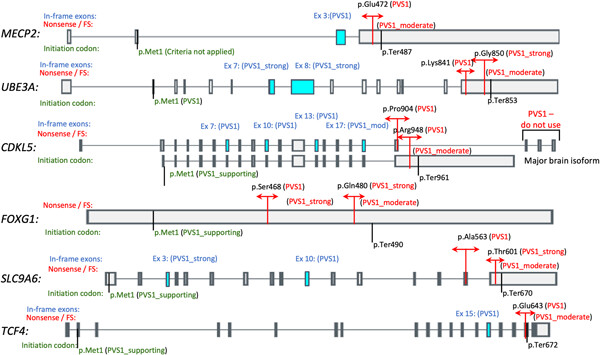
Recommendations by the ClinGen Rett/Angelman-like expert panel for gene-specific variant interpretation methods
- Pages: 1097-1113
- First Published: 27 November 2021
Harmonizing variant classification for return of results in the All of Us Research Program
- Pages: 1114-1121
- First Published: 19 December 2021