Journal list menu
Export Citations
Download PDFs
Table of Contents
Development of Broad-Spectrum Halomethyl Ketone Inhibitors Against Coronavirus Main Protease 3CLpro
- First Published: 28 June 2008
Peptide Nanoparticles as Novel Immunogens: Design and Analysis of a Prototypic Severe Acute Respiratory Syndrome Vaccine
- First Published: 18 December 2008
In Silico Prediction of SARS Protease Inhibitors by Virtual High Throughput Screening
- First Published: 24 April 2007
Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors
- First Published: 22 August 2019
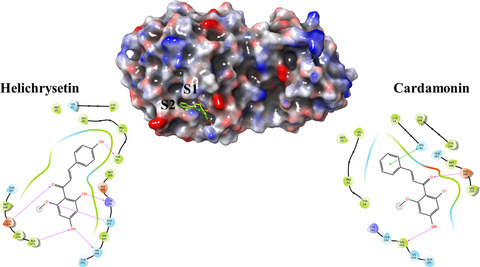
MERS-CoV 3CLpro was assayed with a flavonoid library and some chalcone derivatives represented promising inhibitory activity. The docking study confirmed the experimental measurement of the better efficacy of helichrysetin than cardamonin. This study showed the antiviral effect of flavonoids in the atomic level.
Important Changes in Biochemical Properties and Function of Mutated LLP12 Domain of HIV-1 gp41
- First Published: 10 September 2007
AVCpred: an integrated web server for prediction and design of antiviral compounds
- First Published: 04 August 2016
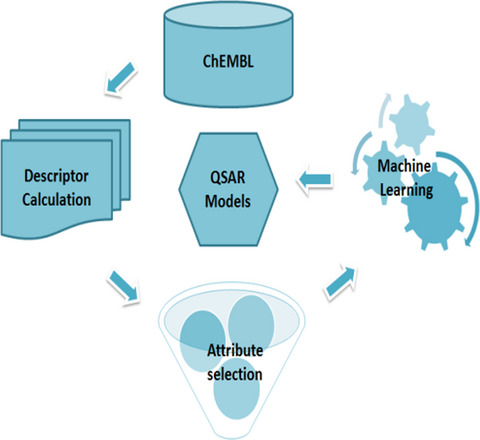
AVCpred is a QSAR approach to predict the antiviral potential of chemical compounds using relationships connecting molecular descriptors and inhibition. Experimentally validated antiviral compounds against important human viruses from ChEMBL database were used to develop the prediction models. The web server with user friendly prediction/design options also has other tools for analysis.
Synthetic peptides from the N-domains of CEACAMs activate neutrophils
- First Published: 08 December 2008




