Journal list menu
Export Citations
Download PDFs
editorial
Using resources generated by the Seattle Structural Genomics Center for Infectious Disease (SSGCID) for training early career researchers
- Pages: 222-225
- First Published: 14 May 2025

The focused issue on Empowering education through structural genomics is introduced. The virtual issue is available at https://journals.iucr.org/special_issues/2024/educationsg.
research communications
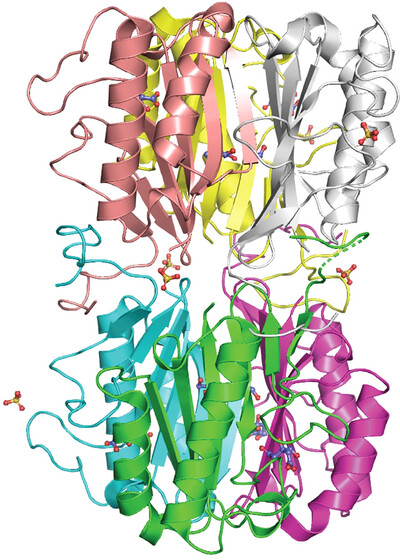
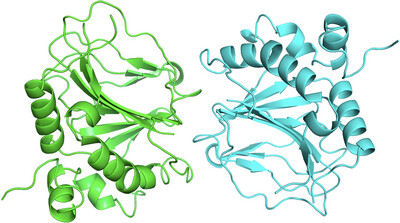
Crystal structures of the putative endoribonuclease L-PSP from Entamoeba histolytica
- Pages: 226-234
- First Published: 14 May 2025
Structure of an Fe2+-binding-deficient mimiviral collagen lysyl hydroxylase
- Pages: 235-240
- First Published: 14 May 2025
Crystal structure of a recombinant Agaricus bisporus mushroom mannose-binding protein with a longer C-terminal region
- Pages: 241-248
- First Published: 14 May 2025
Crystal structure of ATP-dependent DNA ligase from Rhizobium phage vB_RleM_P10VF
- Pages: 249-254
- First Published: 14 May 2025
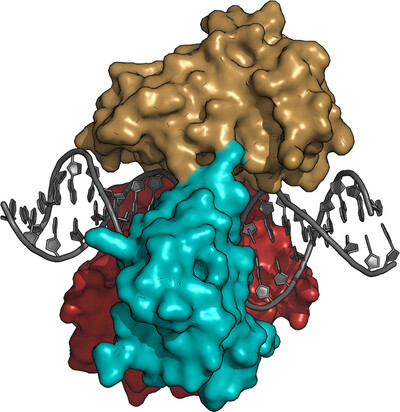
We have determined the structure of the Rhizobium phage vB_RleM_P10VF DNA ligase bound to a nicked DNA duplex to 2.2 Å resolution. The DNA ligase exhibits a canonical DNA ligase-binding mode fully encircling the duplex and has considerable structural homology to T4 DNA ligase and the bacterial ATP-dependent DNA ligase from Prochlorococcus marinus.
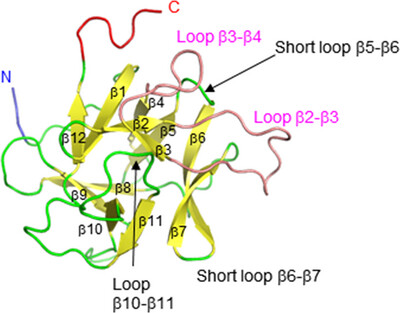
Crystal structure of the C1 domain of the surface-layer protein SlpM from Lactobacillus brevis: a module involved in protein self-assembly
- Pages: 255-262
- First Published: 19 May 2025
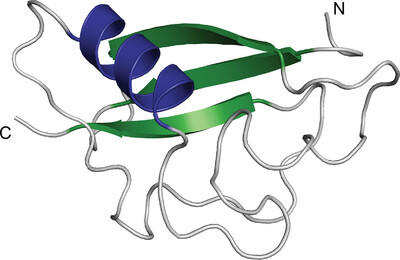
Structural analysis of YcdY, a member of the redox-enzyme maturation protein family
- Pages: 263-271
- First Published: 19 May 2025
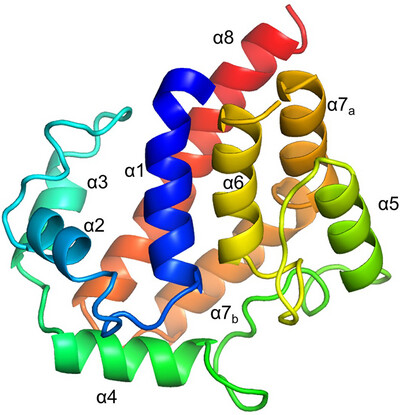
YcdY, a putative chaperone of the NarJ subfamily, forms a helix-bundle structure characterized by a dent on the concave side. The dent contains hydrophobic or conserved residues, presumably for the chaperone function of YcdY. Furthermore, we propose that YcdY may function as a chaperone for proteins other than the previously proposed YcdX.