Journal list menu
Export Citations
Download PDFs
Cover, Volume 43, Issue 6
- Page: i
- First Published: 20 May 2022

Front Cover: The cover image is based on the Editorial Seven years since the launch of the Matchmaker Exchange: the evolution of genomic matchmaking by Heidi L Rehm et al., https://doi.org/10.1002/humu.24373.
Seven years since the launch of the Matchmaker Exchange: The evolution of genomic matchmaking
- Pages: 659-667
- First Published: 10 May 2022
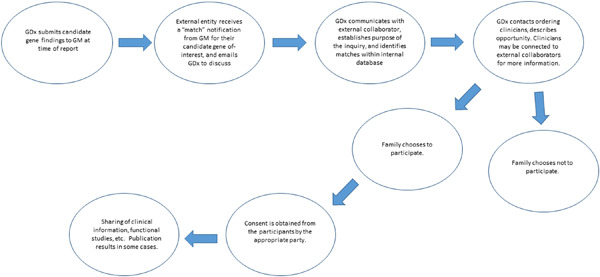
The impact of GeneMatcher on international data sharing and collaboration
- Pages: 668-673
- First Published: 16 February 2022
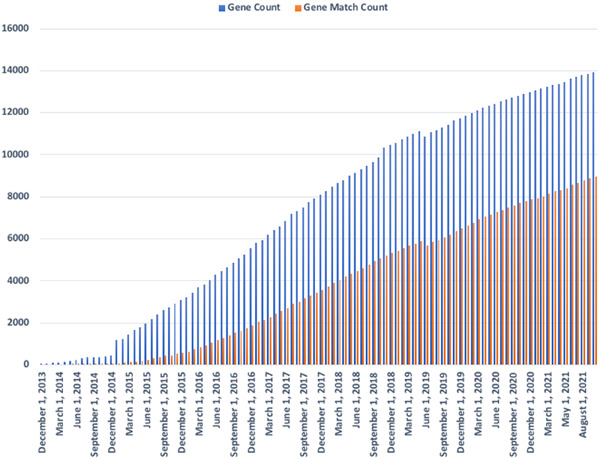
GeneMatcher is the leading resource for connecting individuals with an interest in the same gene from around the world to create collaborations and generate the evidence needed to support novel disease gene identification. GeneMatcher is a founding member of the Matchmaker Exchange (matchmakerexchange.org) and all GeneMatcher submissions are open to matching from any connected node. The thousands of unique gene matches have already generated over 500 publications.
PhenomeCentral: 7 years of rare disease matchmaking
- Pages: 674-681
- First Published: 14 February 2022
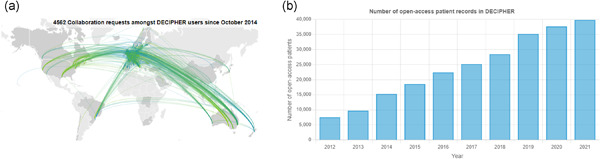
DECIPHER: Supporting the interpretation and sharing of rare disease phenotype-linked variant data to advance diagnosis and research
- Pages: 682-697
- First Published: 10 February 2022
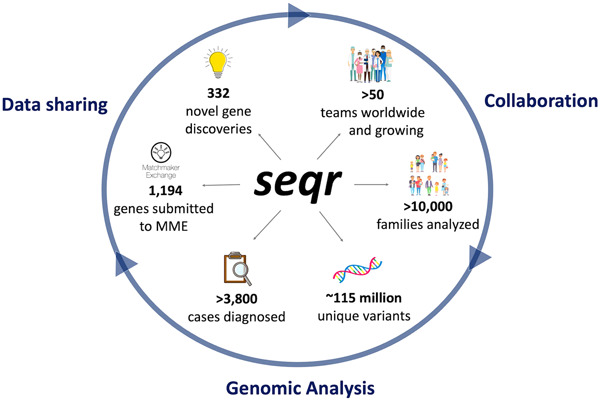
seqr: A web-based analysis and collaboration tool for rare disease genomics
- Pages: 698-707
- First Published: 09 March 2022
PatientMatcher: A customizable Python-based open-source tool for matching undiagnosed rare disease patients via the Matchmaker Exchange network
- Pages: 708-716
- First Published: 22 February 2022
The RD-Connect Genome-Phenome Analysis Platform: Accelerating diagnosis, research, and gene discovery for rare diseases
- Pages: 717-733
- First Published: 17 February 2022
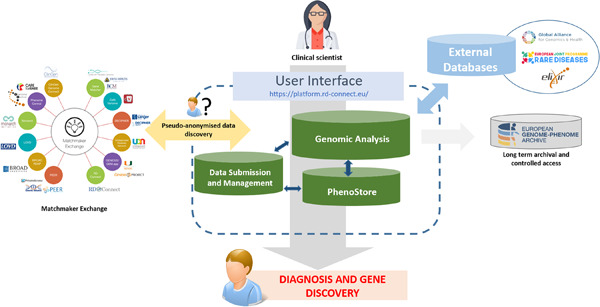
The RD-Connect Genome-Phenome Analysis Platform (GPAP) is a scalable and interoperable online system which facilitates the collation, analysis, interpretation and sharing of integrated genome-phenome datasets, with a particular focus on RD case diagnosis and novel gene discovery. It is free to use for all noncommercial members of the rare disease research community.
Advances in the development of PubCaseFinder, including the new application programming interface and matching algorithm
- Pages: 734-742
- First Published: 10 February 2022
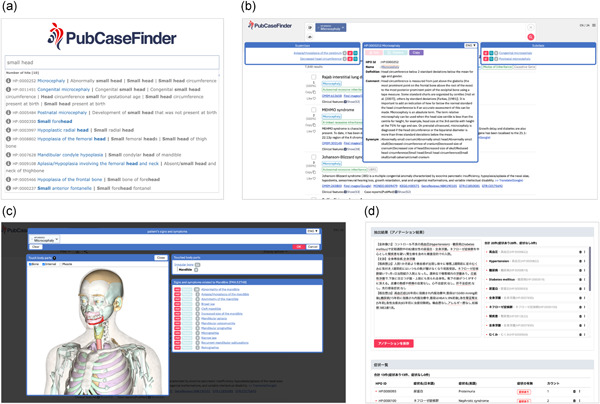
In September 2017, we released PubCaseFinder (https://pubcasefinder.dbcls.jp), a web-based clinical decision support system that provides ranked lists of genetic and rare diseases using Human Phenotype Ontology-based phenotypic similarities, where top-listed diseases represent the most likely differential diagnosis. In this paper, we describe notable updates regarding PubCaseFinder, the GeneYenta matching algorithm implemented in PubCaseFinder, and the PubCaseFinder API.
ModelMatcher: A scientist-centric online platform to facilitate collaborations between stakeholders of rare and undiagnosed disease research
- Pages: 743-759
- First Published: 27 February 2022
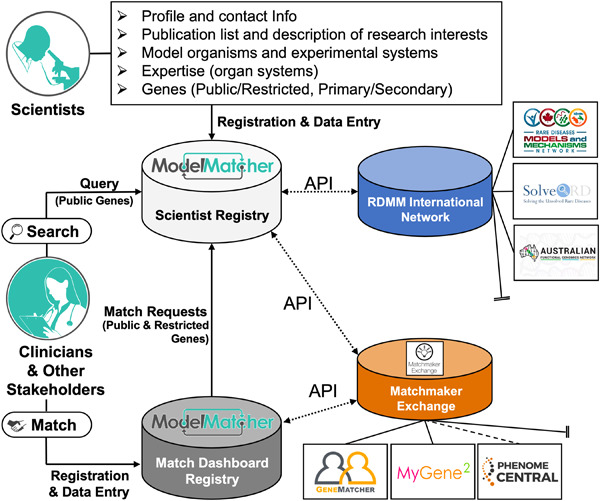
ModelMatcher is a matchmaking website that connects various stakeholders of rare and undiagnosed research to scientists with specific expertise and interests. This platform is connected with large clinical and scientific registries through Matchmaker Exchange and Rare Diseases Models and Mechanisms Networks, respectively, facilitating cross-disciplinary collaborations on a global scale.
Discovery of over 200 new and expanded genetic conditions using GeneMatcher
- Pages: 760-764
- First Published: 16 February 2022
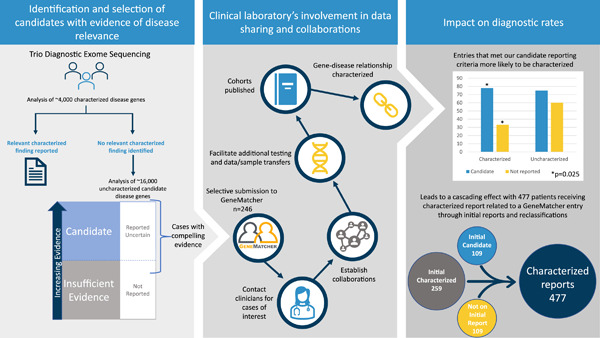
A clinical laboratory's experience using GeneMatcher—Building stronger gene–disease relationships
- Pages: 765-771
- First Published: 19 February 2022
Diagnostic testing laboratories are valuable partners for disease gene discovery: 5-year experience with GeneMatcher
- Pages: 772-781
- First Published: 10 February 2022
Variant-level matching for diagnosis and discovery: Challenges and opportunities
- Pages: 782-790
- First Published: 22 February 2022
Beacon v2 and Beacon networks: A “lingua franca” for federated data discovery in biomedical genomics, and beyond
- Pages: 791-799
- First Published: 17 March 2022
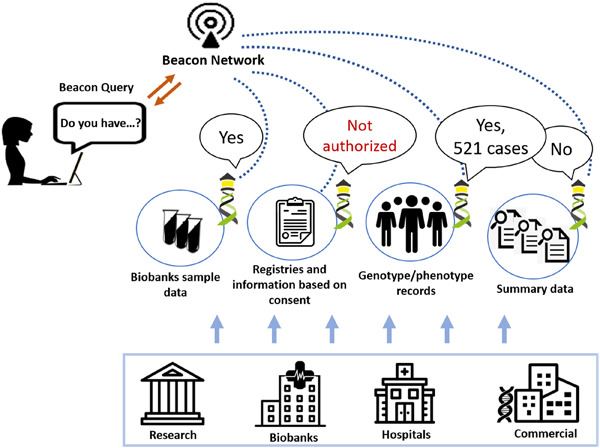
Beacon is a basic data discovery protocol issued by the Global Alliance for Genomics and Health (GA4GH). Beacon version 2 serves real world requirements and addresses the needs of clinical genomics research and healthcare, allowing implementing consortia to return matches in beacon responses and provide a handover to their preferred data exchange format. Beacon is designed as a “lingua franca” to bridge data collections hosted in software solutions with different and rich interfaces. Beacon queries could be send to Beacon instances directly or via Beacon networks. The response could be yes/no, counts or details if the user is properly authorized.
Genomics4RD: An integrated platform to share Canadian deep-phenotype and multiomic data for international rare disease gene discovery
- Pages: 800-811
- First Published: 19 February 2022
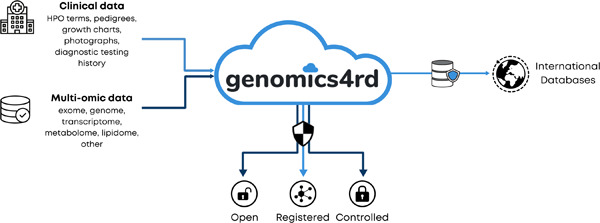
Here we present Genomics4RD, an integrated and internationally accessible web platform, to share, store, and visualize phenotypic and multiomic data from rare disease patients across Canada. Genomics4RD is connected to several other rare disease databases for the purposes of matchmaking and disease-gene discovery.