Journal list menu
Export Citations
Download PDFs
Cover Image, Volume 37, Issue 12
- Page: i
- First Published: 09 November 2016

On the cover: With the rapid growth of NGS applications, we have devoted a full Human Mutation Special Issue to this topic. This unique edition addresses all aspects of the technology and its application, ranging from data generation/variant detection and interpretation to applications in the clinical context and legal/ethical aspects. Future technological developments are also discussed. See the Overview by Madhuri Hegde et al., Page 1247, DOI: 10.1002/humu.23142.
Issue Information
- Pages: 1243-1246
- First Published: 09 November 2016
High-Throughput Sequencing in the Context of Human Genetic Diseases: Now and Tomorrow
- Page: 1247
- First Published: 09 November 2016
Content Analysis of Informed Consent for Whole Genome Sequencing Offered by Direct-to-Consumer Genetic Testing Companies
- Pages: 1248-1256
- First Published: 05 October 2016

WGS can be purchased from commercial companies advertising directly to consumers on the Internet. When purchasing such tests, consumers agree to documents which may not provide adequate information for informed consent as outlined in guidelines for clinical WGS. Among others, issues such as pre-test counselling, the explanation of benefits and risks, as well as the management of incidental findings should be properly addressed during the consent process.
Am I My Family's Keeper? Disclosure Dilemmas in Next-Generation Sequencing
- Pages: 1257-1262
- First Published: 20 September 2016

Whose responsibility is it to warn family members when NGS reveals a life-threatening mutation? Ethical analysis points out that professionals and patients are both responsible and need to work together. Genetic professionals may emphasize the moral obligations of patients using a moral accountability nudge. This strategy is an ethically sound and practically feasible approach for dealing with unsolicited NGS findings.
From Wet-Lab to Variations: Concordance and Speed of Bioinformatics Pipelines for Whole Genome and Whole Exome Sequencing
- Pages: 1263-1271
- First Published: 08 September 2016
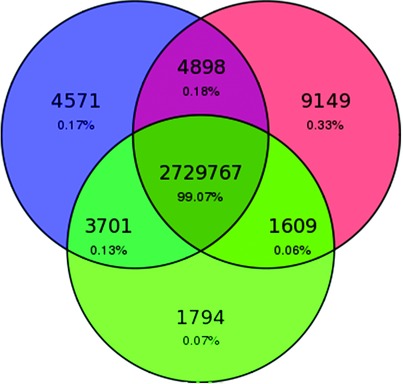
As Whole Genome sequencing becomes cheaper and faster it will progressively substitute targeted sequencing. However, computing cost-performance ratio is not advancing at an equivalent rate. We have benchmarked 6 combinations of state-of-the-art read aligners and variant callers on WGS and WES data. We report differences in speed of up to 20 times in some processes and have observed that SNV, and to a lesser extent InDel, detection is highly consistent only in 70% of the genome.
How to Identify Pathogenic Mutations among All Those Variations: Variant Annotation and Filtration in the Genome Sequencing Era
- Pages: 1272-1282
- First Published: 07 September 2016
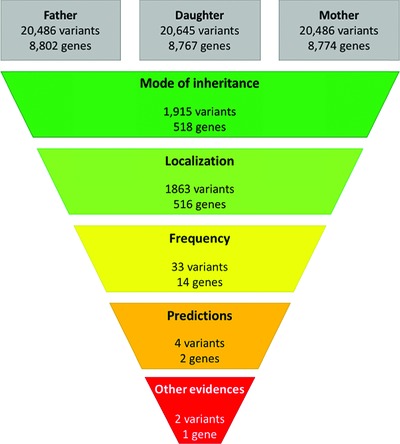
High-throughput sequencing technologies have become fundamental to identify disease-causing mutations in human genetic diseases both in research and clinical testing contexts. However, due to several factors including wrong variant annotation and non-optimal filtration practices some of these mutations can be misinterpreted or overlooked in the pool of detected mutations. In these review we describe filtration options, provide a generic filtration workflow and highlight potential pitfalls through a use case to facilitate identification of disease-causing mutations.
Noncoding RNA: Current Deep Sequencing Data Analysis Approaches and Challenges
- Pages: 1283-1298
- First Published: 12 August 2016
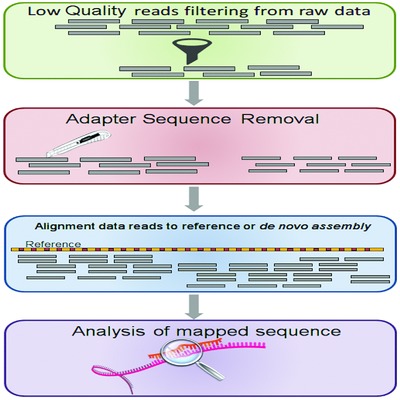
Next Generation Sequencing allows to obtain a more accurate profiling of ncRNA classes, along with a low experimental complexity, concurrently posing an increasing challenge in terms of efficiency, accuracy, and usability of data analysis software. Recent/popular computational approaches generally filter and prepare data similarly, while applying distinct strategies in the alignment and analysis of sequenced reads, in the attempt to attain a more sensitive and comprehensive ncRNA annotation capable of furthering the understanding of this still vastly uncharted genomic territory.
Actionable Genes, Core Databases, and Locus-Specific Databases
- Pages: 1299-1307
- First Published: 07 September 2016

Adoption of next generation sequencing in a diagnostic context raise numerous questions regarding identification and reporting of secondary variants (SVs) in actionable genes. Here, we wondered if Locus Specific databases (LSDBs) could advance the interpretation of missense variants as they can provide a wealth of information and enable classifying variants. Thanks to a use case, we demonstrated LSDBs' benefits to annotate SVs and minimize over interpretation of mutations because of their efficient curation process and collection of unpublished data.
WES/WGS Reporting of Mutations from Cardiovascular “Actionable” Genes in Clinical Practice: A Key Role for UMD Knowledgebases in the Era of Big Databases
- Pages: 1308-1317
- First Published: 20 September 2016

Next-generation sequencing leads to the identification of secondary variants. Among the 56 “actionable” genes recommended by the ACMG for their reporting in clinical practice, seven are involved in Marfan syndrome and related disorders: FBN1, TGFBR1, TGFBR2, ACTA2, SMAD3, MYH11 and MYLK. Here, we show that UMD Locus Specific Databases report mutations rarely found in global scale initiatives, provide access to full spectrum of known pathogenic mutations and are then the best resource to interpret primary and secondary variants.
BRCA Share: A Collection of Clinical BRCA Gene Variants
- Pages: 1318-1328
- First Published: 16 September 2016
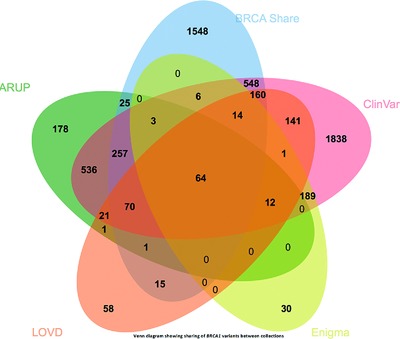
BRCA ShareTM is an international alliance of academic centers and commercial testing laboratories. By integrating content of the Universal Mutation Database generated by the French Unicancer Genetic Group with the testing results of Quest Diagnostics and Laboratory Corporation of America (LabCorp), BRCA ShareTM has assembled one of the largest publicly accessible collections of BRCA variants with over 6,200 variants currently available. In addition, to funding high quality curation, BRCA ShareTM seeks to support functional studies, targeting variants of uncertain significance.
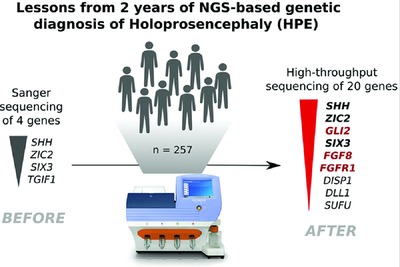
Mutational Spectrum in Holoprosencephaly Shows That FGF is a New Major Signaling Pathway
- Pages: 1329-1339
- First Published: 01 July 2016
Mini-Exome Coupled to Read-Depth Based Copy Number Variation Analysis in Patients with Inherited Ataxias
- Pages: 1340-1353
- First Published: 16 August 2016
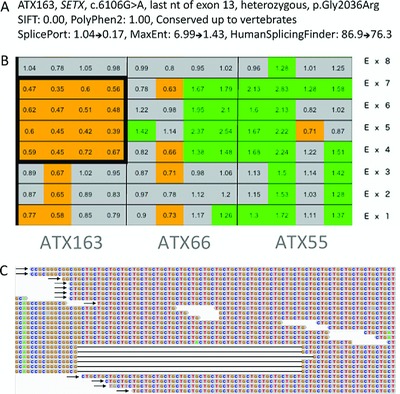
Identification of nucleotide variations, copy number variations, and expansion in inherited ataxias by next generation sequencing:
- – Pathogenicity prediction of nucleotide (nt) variation (Figure A): multiscoring predicts both missense and splice site pathogenicity for the same heterozygous SETX variation in patient ATX163
- – CNV detection (Figure B): heterozygous SETX exon 4 to 7 deletion in patient ATX163
- – Expansion detection (Figure C): ATXN2, CAG expansion (arrows) in patient ATX340
An Application of NGS for Molecular Investigations in Perrault Syndrome: Study of 14 Families and Review of the Literature
- Pages: 1354-1362
- First Published: 21 September 2016
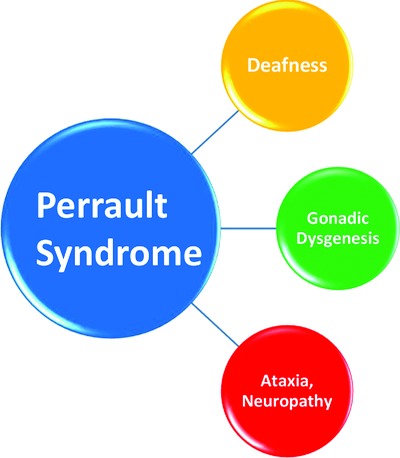
Perrault syndrome is a rare autosomal recessive condition characterized by deafness and gonadic dysgenesis. In the literature 98 cases have been reported and only 25 mutations in the five Perrault genes have been identified. Thanks to NGS, four novel mutations have been identified in four Perrault genes (C10orf2, CLPP, HARS2 and LARS2). Molecular analysis confirmed the clinical diagnosis in 28.6% and some genotype-phenotype correlations were established.
Fourth Generation of Next-Generation Sequencing Technologies: Promise and Consequences
- Pages: 1363-1367
- First Published: 13 July 2016
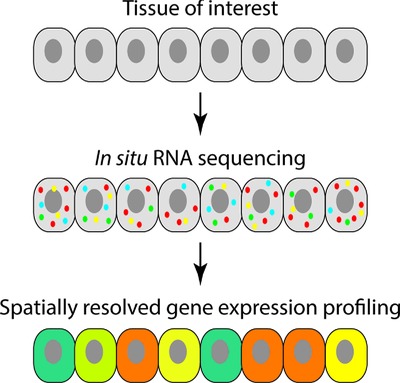
In this review, we discuss the emergence of 4th generation sequencing technologies that provides RNA and DNA sequences with their exact spatial localization preserved within cells or tissue. These new technologies open up new opportunities for studying gene expression and their regulatory networks within tissue with cellular resolution.