WES/WGS Reporting of Mutations from Cardiovascular “Actionable” Genes in Clinical Practice: A Key Role for UMD Knowledgebases in the Era of Big Databases
Contract grant sponsor: Aix-Marseille Université; Contract grant sponsor: INSERM; Contract grant sponsor: The European Union Seventh Framework Program; Grant number: 305444.
For the Next Generation Sequencing special issue
ABSTRACT
High-throughput next-generation sequencing such as whole-exome and whole-genome sequencing are being rapidly integrated into clinical practice. The use of these techniques leads to the identification of secondary variants for which decisions about the reporting or not to the patient need to be made. The American College of Medical Genetics and Genomics recently published recommendations for the reporting of these variants in clinical practice for 56 “actionable” genes. Among these, seven are involved in Marfan Syndrome And Related Disorders (MSARD) resulting from mutations of the FBN1, TGFBR1 and 2, ACTA2, SMAD3, MYH11 and MYLK genes. Here, we show that mutations collected in UMD databases for MSARD genes (UMD-MSARD) are rarely reported, including the most frequent ones, in global scale initiatives for variant annotation such as the NHLBI GO Exome Sequencing Project (ESP), the Exome Aggregation Consortium (ExAC), and ClinVar. The predicted pathogenic mutations reported in global scale initiatives but absent in locus-specific databases (LSDBs) mainly correspond to rare events. UMD-MSARD databases are therefore the only resources providing access to the full spectrum of known pathogenic mutations. They are the most comprehensive resources for clinicians and geneticists to interpret MSARD-related variations not only primary variants but also secondary variants.
Introduction
Since 2005, Next-Generation DNA Sequencing (NGS) platforms have been implemented largely leading to reduced DNA sequencing cost by four orders of magnitude relative to Sanger sequencing. Consequently, clinical use of Whole-Exome Sequencing (WES) and Whole-Genome Sequencing (WGS) is increasing [Institute of Medicine (US) 2012]. This cost-effective option is becoming the technique of choice in the work-up of disorders that involve multiple genes and proves its effectiveness in the identification of genes involved in previously undiagnosed cases (near 25%) [Yang et al., 2013], or in a specifically targeted region [Bamshad et al., 2011]. Thus, NGS applications to new medical situations are emerging including personalized treatment (notably for cancer) [Cotterell, 2014; Sun and Califano, 2014], pharmacogenomics [Harper and Topol, 2012], preconception/prenatal screening [Fan et al. 2012; Carss et al. 2014], or population screening for disease risk [Biesecker, 2012]. Nevertheless, these technologies generate ethical issues with the identification of variants in population that may be pathogenic, called "secondary variants" (previously called “incidental findings”), unrelated to the indication for ordering the sequencing but of medical value for patient care [Christenhusz et al., 2013]. The topic is controversial, highlighting the need of optimized informed consent procedure [Rigter et al., 2013], but also clinicians’ obligations or not to identify and disclose such findings [Biesecker, 2013; Clayton et al., 2013]. Although developing consensus in the major part of Europe is to use targeted approaches or to limit the analysis to specific sets of genes in order to avoid unsolicited findings [van El et al., 2013], the American College of Medical Genetics and Genomics (ACMG) issued recommendations for reporting these “secondary variants” in clinical practice [Green et al., 2013]. They recommend the reporting of all pathogenic mutations, irrespective of patient age, for a specific set of 56 genes associated with 24 highly penetrant inherited conditions. Among these genes, seven are involved in Marfan Syndrome And Related Disorders (MSARD) (including familial Thoracic Aortic Aneurysms and Dissections (fTAAD), Aortic Osteoarthritis Syndrome (AOS), and Loeys-Dietz syndrome): FBN1 (MIM* 134797), TGFBR1 (MIM* 190181) and TGFBR2 (MIM* 190182), ACTA2 (MIM* 102620), SMAD3 (MIM* 603109), MYH11 (MIM* 160745), and MYLK (MIM* 600922) genes.
WGS/WES technologies generate a tremendous amount of data. As evaluation of variations to identify pathogenic mutations is labor intensive, very rare or novel changes are distinguished by filtering against a set of variants that are available in public databases such as dbSNP [Sherry et al., 2001], 1000 Genomes Project [1000 Genomes Project Consortium et al., 2015], Exome Sequencing Project (ESP), and UK 100K genome or Exome Aggregation Consortium (ExAC) [Lek et al., 2016]. This filtering can eliminate truly pathogenic mutations already reported in these sets as there is today no way to identify the phenotype associated with variations in individuals from these databases. Consequently, a filter chain that removes variations according to a too low Minor Allele Frequency (MAF) could lead to misinterpretation.
Locus-Specific Databases (LSDB) are today essential as more and more diagnostic laboratories worldwide are using NGS technologies without specific expertise for each of these 56 genes. Access to the full spectrum of already known pathogenic mutations, as well as combined interpreted data from many reference diagnostic laboratories, can help clinicians and geneticists in interpreting variants. They can rely on these reference databases to rapidly collect relevant information for data interpretation, report more accurate conclusions and save time. They will also be able to answer the following questions: “Has this variant already been described in others patients?”, “Which phenotype is associated with this mutation?”, “What are the predictions and evidences for its pathogenicity?”, “Is the mutation associated with cardiovascular risk?”. This last information is needed to apply the last nosology for Marfan syndrome (Ghent 2 nosology) [Loeys et al., 2010; Faivre et al., 2012].
Materiel and Methods
UMD Databases
In an effort to standardize information regarding mutations in the FBN1 gene, we developed in 1995 a locus-specific database [Collod et al., 1996; Collod-Beroud et al., 2003] with the generic system called Universal Mutation Database (UMD) [Béroud et al., 2000, 2005]. Subsequently, a database for TGFBR2 [Frédéric et al., 2008] gene mutations was created. To be exhaustive and facilitate NGS analysis, five other databases for TGFBR1, ACTA2, SMAD3, MYH11, and MYLK genes have been developed since 2012. They contain all known pathogenic mutations collected from literature and through direct collaborations with diagnostic laboratories as well as some polymorphisms. Relatives have been implemented in databases at the end of 2014 for TGFBR1 and 2, SMAD3, ACTA2, MYH11, and MYLK genes and this work is in progress for the UMD-FBN1 database. Each variation is annotated at the gene (exon and codon number, wild-type and mutant codons), protein (wild-type and mutant amino acids, highly conserved domain), and clinical levels (clinical signs identified in the patient, when available). The UMD databases are updated and curated by experts. All these databases are accessible at: http://www.umd.be/.
UMD-MSARD Data Extraction
UMD-MSARD databases contain to date 3,315 entries for the FBN1 gene, and 130, 213, 61, 209, 45, and 13 for TGFBR1, TGFBR2, SMAD3, ACTA2, MYH11, and MYLK genes, respectively.
In order to constitute the list of mutations to be compared, all polymorphisms described in UMD databases have been excluded and only different pathogenic mutations from probands have been selected (corresponding to the number of different mutational events or unique variants). Data extraction lead to a list of 1,976 different mutational events for the FBN1 gene and, respectively, 46, 119, 15, 39, 10, and 5 mutational events for TGFBR1, TGFBR2, SMAD3, ACTA2, MYH11, and MYLK genes, respectively.
Data Extraction from ExAC, ESP, and Clinvar
We searched for all variations matching reference transcripts (FBN1: ENST00000316623, TGFBR1: ENST00000374994, TGFBR2: ENST00000295754, SMAD3: ENST00000327367, ACTA2: ENST00000224784, MYH11: ENST00000300036, and MYLK: ENST00000360304) from Exome Aggregation Consortium (ExAC) Cambridge, MA (http://exac.broadinstitute.org) [Lek et al., 2016]. Variants from the NHLBI GO Exome Sequencing Project (ESP) were extracted from the file provided by the Annovar Tool [Wang et al., 2010] as well as information from Ensembl (GRCh release 75) and Clinvar (September 29, 2014) (http://www.ncbi.nlm.nih.gov/clinvar/).
To select potential pathogenic mutations in ExAC variations matching reference transcripts, we applied the UMD-Predictor® tool (http://www.umd-predictor.eu) [Frédéric et al., 2009; Salgado et al., 2016] to predict the pathogenicity of missense variations. We had previously evaluated its accuracy and performances relatively to the seven most used and reliable prediction tools (SIFT 5.1.1 [Sim et al., 2012], Polyphen 2.2.2 [Adzhubei et al., 2013], Provean 1.1.3 [Choi et al., 2012], Mutation Assessor 2 [Reva et al., 2011], CONDEL 1.5 [González-Pérez and López-Bigas, 2011], MutationTaster 2 [Schwarz et al., 2014], and CADD [Kircher et al., 2014]). The largest reference variation datasets including more than 140,000 annotated variations (Varibench [Sasidharan Nair and Vihinen, 2013] with dbSNP [Sherry et al., 2001], UniProt [UniProt Consortium, 2014], Clinvar [Landrum et al., 2016], and PredictSNP [Bendl et al., 2014]) have been used for these tests. UMD-Predictor consistently demonstrated a better accuracy (0.85), specificity (0.95), Matthews correlation coefficient (0.69), and Diagnostic Odds Ratio (86.6) [Salgado et al., 2016]. Nonsynonymous missenses variations with a UMD-Predictor prediction score between 65 and 74 corresponding to “probably pathogenic” variations and with a prediction score superior to 74 corresponding to “pathogenic” variations have been selected. For synonymous missense variations as well as variations located in consensus acceptor, donor, or branch point sites the Human Splicing Finder® tool [Desmet et al., 2009] (http://www.umd.be/HSF3/) have been applied to identify variations with highly probable impact on splicing (WT donor, acceptor, or branch point splice site broken). Variations creating a cryptic donor or acceptor splice site in a favorable environment have also been selected. Frameshift deletions or insertions have been considered as pathogenic. Intronic variations not localized in consensus splice site, in frame deletions or insertions have not been included as the pathogenic character of these variations is unclear without in vitro assays. Finally, variants from ExAC, ESP, and UMD-Databases were merged into one table using a homemade Perl script.
Data extraction for FBN1 Variations Discussed by Yang et al. 2014
Yang et al. (2014) selected previously Marfan-associated variants by combining query in Human Gene Mutation Database (HGMD professional 2013.2) and PudMed. They identified by this way 891 variants among which only 23 have frequencies reported in ESP.
Results
UMD Databases and Other Public Databases (ESP, ExAC, and ClinVar)
The UMD databases, respectively, contain: FBN1 (3,315 entries), TGFBR1 (130 entries), TGFBR2 (213 entries), SMAD3 (61 entries), ACTA2 (209 entries), MYH11 (45 entries), and MYLK (13 entries) (Table 1) and are accessible at: http://www.umd.be/. They correspond to all known pathogenic mutations collected from literature until 2014 and through direct collaborations with diagnostic laboratories. They contain also some variations sometimes published as mutations but subsequently demonstrated or corresponding more likely to nonpathogenic variations (reported in databases as “polymorphism”). The different pathogenic variations described for probands (corresponding to the number of different mutational events or unique variants) are distributed as follow: 1,976 index cases for FBN1, and, respectively, 46, 119, 15, 39, 10, and 5 for TGFBR1, TGFBR2, SMAD3, ACTA2, MYH11, and MYLK genes.
| Gene | FBN1 | TGFBR1 | TGFBR2 | SMAD3 | ACTA2 | MYH11 | MYLK | Total |
|---|---|---|---|---|---|---|---|---|
| Number of entries in UMD databases | 3,315 | 130 | 213 | 61 | 209 | 45 | 13 | |
| Number of different pathogenic mutational events reported in UMD databases | 1,976 | 46 | 119 | 15 | 39 | 10 | 5 | 2,210 |
| Pathogenic mutations reported in ExAC | 67 | 4 | 9 | 0 | 4 | 3 | 1 | 88 |
| Highest allele frequency in ExAC | 3.35 × 10−3 | 2.76 × 10−4 | 1.02 × 10−3 | / | 1.30 10−4 | 7.32 10−4 | 4.40 × 10−4 | |
| Lowest allele frequency in ExAC | 8.13 × 10−6 | 8.13 × 10−6 | 8.13 × 10−6 | / | 8.13 × 10−6 | 1.63 × 10−5 | 6.52 × 10−5 | |
| Pathogenic mutations reported in ESP (ESP variations not found in ExAC) | 28(1) | 1 | 2 | 0 | 0 | 3 | 2 | 36 |
| Highest allele frequency | 1.20 10−2*(0.0019250) | 4.61 × 10−4 | 7.70 × 10−5 | 0 | 0 | 5.40 × 10−4 | 1.54 × 10−4 | |
| Lowest allele frequency | 7.70 10−5 | 4.61 × 10−4 | 7.70 × 10−5 | 0 | 0 | 7.70 × 10−5 | 7.70 × 10−5 | |
| Pathogenic mutations reported in ClinVar | 15 | 1 | 1 | 0 | 0 | 0 | 0 | 17 |
- Highest and lowest allele frequencies found in ExAC and ESP are reported.
- *Highest allele frequency found for the FBN1 variation c.6832C>T (p.Pro2278Ser) is followed by the next highest frequency.
The global molecular analysis of these databases reveals that missense mutations represent more than half of the events (Fig. 1). Being able to distinguish neutral sequence variations from those responsible for the phenotype is of major interest in clinical diagnosis. Since in vitro validation of mutations is not always possible, indirect arguments have to be accumulated to define if a missense variation is causative, beginning with mutation segregation in affected family members or absence in both parents in sporadic cases (de novo cases). In the absence of an adequate functional test, absence of this variation in a panel of at least 300 independent population matched control chromosomes (now low reported frequencies in core databases), biochemical nature of the substitution, protein region where the variation is located, and degree of conservation among species are some of the classical evidences in favor of a pathogenic mutation that reference diagnostic laboratories have to regroup. The collection of these data is often both time consuming and costly. This is quickly overcome if the mutation is already reported in one of the UMD-LSDBs.
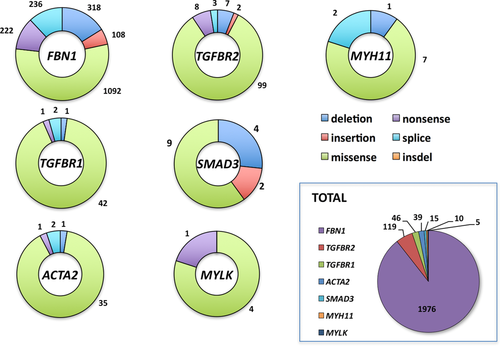
Recently, the release of whole-exome data from the NHLBI GO Exome Sequencing Project (ESP), from the Exome Aggregation Consortium (ExAC) [Lek et al., 2016], or ClinVar [Landrum et al., 2016] open a new source of information to evaluate genetic variation in the general population. In order to evaluate the number of UMD-MSARD variations reported in exome data, we searched for all variations described in ESP, ExAC, and ClinVAr matching reference transcripts for MSARD genes. Then mutational events for each UMD-LSDB [1,976 (FBN1), 46 (TGFBR1), 119 (TGFBR2), 15 (SMAD3), 39 (ACTA2), 10 (MYH11) and 5 (MYLK)] were compared with this list of variations. Among the 2,210 different UMD-MSARD mutational events, 88 variations (4%) have been reported in ExAC, 36 in ESP (1.6%), and 17 in ClinVar (0.7%) (Table 1).
For the diseases associated with these genes, estimated population prevalence ranges between 1:5,000 and 1:4,000.000 in adults depending on the occurrence of an isolated thoracic aortic aneurysm or as a symptom of a syndromic disorder (Arslan-Kirchner et al. 2016). Based on the prevalence of the most common form, Marfan syndrome (1/5,000 = 0.02%), the threshold for allele frequency would be up to 10−4. We checked allele frequencies for all the 88 mutational events reported in our seven UMD-LSDBs and found in ExAC or ESP. Twenty-one missense mutations were found with frequencies higher than the threshold of 10−4 in ExAC (Table 2): 15 variants for FBN1, 2 variants for MYH11, 1 variant for TGFBR1, TGFBR2, ACTA2 and MYLK, respectively. No variant was found for SMAD3 gene.
| Gene | Exon | Mutation name (c.) | Mutation name (p.) | ESP frequencies | ExAC frequencies | Number of probands described with this variation in UMD-LSDBs | Associated mutations, other arguments |
|---|---|---|---|---|---|---|---|
| FBN1 | Exon 2 | c.59A>Ga | p.Tyr20Cys | 0.000231 | 0.0001464 | 1 | FS |
| Exon 10 | c.1027G>Aa | p.Gly343Arg | 0.000154 | 0.0001545 | 1 | ||
| Exon 25 | c.2927G>Aa | p.Arg976His | 0.000154 | 0.0001382 | 2 | ||
| Exon 25 | c.2956G>A | p.Ala986Thr | 0.001309 | 0.001496 | 1 | FS. Cosegregate with c.1001_1073del (trans) FBN1 mutation | |
| Exon 25 | c.3058A>Ga | p.Thr1020Ala | 0.000231 | 0.0004554 | 4 | Cosegregate with a TGFBR1 mutation in 1/4 patient | |
| Exon 28 | c.3422C>Ta | p.Pro1141Leu | 0.001078 | 0.0007237 | 3 | ||
| Exon 29 | c.3509G>Aa | p.Arg1170His | 0.001925 | 0.001163 | 9 | FS in 4/9 patients | |
| Exon 35 | c.4270C>Ga | p.Pro1424Ala | 0.000308 | 0.000187 | 11 | Cosegregate with c.8038C>T FBN1 mutation in 1/11 patient | |
| Exon 36 | c.4441A>G | p.Ser1481Gly | 0.001155 | 0.0003497 | 1 | Cosegregate with c.IVS61+1G>A (c.7699+1G>A) FBN1 mutation | |
| Exon 50 | c.6073G>T | p.Ala2025Ser | 0.001617 | 0.0004554 | 1 | Cosegregate with a SMAD3 mutation | |
| Exon 56 | c.6832C>T | p.Pro2278Ser | 0.011934 | 0.003342 | 2 |
|
|
| Exon 62 | c.7661G>Aa | p.Arg2554Gln | 0.000077 | 0.0001464 | 2 | FS in 1/2 patient | |
| Exon 64 | c.7852G>Aa | p.Gly2618Arg | 0.000154 | 0.0002521 | 3 | 1/3 de novo | |
| Exon 65 | c.8149G>A | p.Glu2717Lys | 0.000077 | 0.0001545 | 2 | Cosegregate with c.3412T>C FBN1 mutation in 1/2 patient | |
| Exon 65 | c.8176C>Ta | p.Arg2726Trp | 0.001078 | 0.0007237 | 16 | FS in 5/16. Cosegregate with FBN1 mutation:
|
|
| TGFBR1 | Exon 9 | c.1433A>G | p.Asn478Ser | 0.000461 | 0.0002765 | 1 | |
| TGFBR2 | Exon 4 | c.944C>T | p.Thr315Met | 0.000077 | 0.001025 | 2 | FS, p.Thr315Met cannot restore growth inhibition in response to TGFß in DR-26 cells |
| ACTA2 | Exon 8 | c.977C>A | p.Thr326Asn | NA | 0.0001301 | 2 | FS |
| MYH11 | Exon 16 | c.2005C>T | p.Arg669Cys | 0.000231 | 0.0005692 | 1 | |
| Exon 33 | c.4673C>T | p.Thr1558Met | 0.000539 | 0.0007319 | 1 | ||
| MYLK | Exon 24 | c.4195G>A | p.Glu1399Lys | 0.000154 | 0.0004391 | 1 |
- FS, cosegregation with disease in the family.
- a Bold variations also discussed by Yang et al. (2014). Nucleotide numbering uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1.
These 15 FBN1 variants are carried by 59 patients among which 10 patients are described as double mutants in UMD-FBN1 (Table 2). They carried a second FBN1 variation predicted pathogenic with UMD-Predictor [Salgado et al., 2016] and with frequencies below 10−4. Two other patients were finally found as carriers of a pathogenic TGFBR1 or a SMAD3 mutation, respectively, with adequate ExAC frequencies. These results suggest that, for these 12 patients, FBN1 variations with frequencies above 10−4 in ExAC may not be the cause of the disease for these patients but very rare polymorphisms, potentially with a modifying effect, cosegregating with the disease in these families. More investigations have to be made in the remaining 49 patients to identify the potential pathogenic variants. Nevertheless, only functional evidences could validate these hypotheses.
These results can be compared with the analyses of Yang et al. [2014] (Table 3). These authors highlighted the little knowledge regarding distribution of mutations in the general population at the time of mutation identification leading to potential false-positive findings. They extracted from HGMD and PubMed 891 previously MFS-associated FBN1 variants in order to compare their ESP frequencies with frequencies expected according to the phenotype prevalence in the general population. Only 23/891 FBN1 variants were described in ESP. Yang et al. postulated that the expected prevalence of MFS in the ESP population is 0.02% (95% CI 0.0%-0.05%) that is 1.3 carriers out of 6,503 subjects. Therefore, the estimated number of individuals affected by MFS in the ESP can be expected to be no more than two. With this conservative approach, 10 of the 23 selected variants were present in three or more individuals in the ESP population and could be considered according to the authors as not being the monogenic cause of MFS but rather as rare polymorphisms (Table 3, variations indicated with an *).
| Mutation name | Nomenclature (protein) | Yang's status | Status in UMD-FBN1 | UMD-Predictor score | ESP frequencies | ExAC frequencies |
|---|---|---|---|---|---|---|
| Rare polymorphisms (Yang's status in accordance with ExAC frequencies) | ||||||
| c.59A>Ga | p.Tyr20Cys | Rare polymorphism | Mutation | 78 (Pathogenic) | 0.000231 | 0.000146a |
| c.3058A>Ga | p.Thr1020Ala | Rare polymorphism | Mutation | 75 (Pathogenic) | 0.000231 | 0.000455a |
| c.3422C>Ta | p.Pro1141Leu | Rare polymorphism | Mutation | 84 (Pathogenic) | 0.001078 | 0.000724a |
| c.3509G>Aa | p.Arg1170His | Rare polymorphism | Mutation | 66 (Probably pathogenic) | 0.001925 | 0.001163a |
| c.4270C>Ga | p.Pro1424Ala | Rare polymorphism | Mutation | 90 (Pathogenic) | 0.000308 | 0.000187a |
| c.6700G>Aa | p.Val2234Met | Rare polymorphism | Polymorphism | 24 (Polymorphism) | 0.000616 | 0.000789a |
| c.8176C>Ta | p.Arg2726Trp | Rare polymorphism | Mutation | 68 (Probably pathogenic) | 0.001078 | 0.000724a |
| Rare polymorphisms (Yang's status not in accordance with ExAC frequencies) | ||||||
| c.1027G>A | p.Gly343Arg | Mutation | Mutation | 100 (Pathogenic) | 0.000154 | 0.000155a |
| c.2927G>A | p.Arg976His | Mutation | Mutation | 78 (Pathogenic) | 0.000154 | 0.000138a |
| c.7661G>A | p.Arg2554Gln | Mutation | Mutation | 84 (Pathogenic) | 0.000077 | 0.000146a |
| c.7852G>A | p.Gly2618Arg | Mutation | Mutation | 100 (Pathogenic) | 0.000154 | 0.000252a |
| Mutations (Yang's status in accordance with ExAC frequencies) | ||||||
| c.2056G>A | p.Ala686Thr | Mutation | Mutation | 90 (Pathogenic) | 0.000077 | 0.000081 |
| c.7241G>A | p.Arg2414Gln | Mutation | Mutation | 74 (Pathogenic) | 0.000077 | 0.000024 |
| c.1345G>A | p.Val449Ile | Mutation | Polymorphism | 48 (Polymorphism) | 0.000154 | 0.000057 |
| c.7660C>T | p.Arg2554Trp | Mutation | Mutation | 100 (Pathogenic) | 0.000077 | 0.000016 |
| c.7702G>A | p.Val2568Met | Mutation | Mutation | 81 (Pathogenic) | 0.000077 | 0.000016 |
| c.8081G>A | p.Arg2694Gln | Mutation | Mutation | 66 (Probably pathogenic) | 0.000077 | 0.000008 |
| c.8494A>G | p.Ser2832Gly | Mutation | Mutation | 100 (Pathogenic) | 0.000077 | 0.000008 |
| c.6055G>A | p.Glu2019Lys | Mutation | Mutation | 84 (Pathogenic) | 0.000077 | NA |
| c.7379A>G | p.Lys2460Arg | Mutation | Mutation | 69 (Probably pathogenic) | 0.000154 | 0.000075 |
| Mutations (Yang's status not in accordance with ExAC frequencies) | ||||||
| c.3797A>Ta | p.Tyr1266Phe | Rare polymorphism | Mutation | 72 (Probably pathogenic) | 0.000308 | 0.000098 |
| c.3845A>Ga | p.Asn1282Ser | Rare polymorphism | Mutation | 81 (Pathogenic) | 0.000231 | 0.000073 |
| c.7846A>Ga | p.Ile2616Val | Rare polymorphism | Mutation | 72 (Probably pathogenic) | 0.000308 | 0.000065 |
- The 23 variations described as mutations and reported in ESP have been checked for their status in the UMD-FBN1 database and their frequencies in ExAC.
- a Mutations considered as rare polymorphisms by these authors according to ESP frequencies. Bold: reported as polymorphism in UMD-FBN1. (Nucleotide numbering uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1).
We first compared ESP allele frequencies reported for these 23 mutations with ExAC frequencies (Table 3). Among the ten variants classified as rare plymorphisms (noncausal) by Yang et al., seven (c.59A>G, c.3058A>G, c.3422C>T, c.3509G>A, c.4270C>G, c.6700G>A and 8176C>T) have frequencies in ExAC above 10−4 (Table 3) supporting a “rare polymorphism” status. However, this status was not confirmed for the remaining three variants (c.3797A>T, c.3845A>G and c.7846A>G) as their frequencies were all lower than 10−4 in the ExAC population contrary to ESP. These variants would then be no longer classified as rare polymorphisms according to their frequencies. This discrepancy brings forward the importance of tested population size (6,503 patients in ESP and 60,706 patients in ExAC) and also for potential confounding effects, as reference databases may vary according to their ascertainment procedures. For instance, ESP database is based on numerous projects aiming to decipher Mendelian bases of genetic diseases including cardiovascular disorders (Supp. Table S1). These populations are then potentially not representative of global population because of enrichment for specific clinical conditions possibly leading to overestimated frequencies for some variants. Evaluation of the effect of potential bias of such selection on allele frequency should then be a prerequisite to adequately use these core databases to sort variants according to the expected allele frequency in the general population. Each identified variation is unfortunately not linked to a specific sample nor associated with a specific disease for evident patient confidentiality. Therefore, we were unable to approximate any a priori selection bias.
Frequency of Secondary Variants in MSARD Genes
In order to know how frequent are the secondary variants in MSARD genes, all variations described in ExAC and ESP matching the seven reference transcripts have been annotated with ANOVAR as: exonic (missense, STOP gained, frameshift or inframe insertion/deletion, or synonymous), intronic (deep intronic, ncRNA, or splice regions), 5’ and 3’ UTR. Comparison of variations identified in ESP (Table 4) and ExAC (Table 5) showed that the large majority of ESP variations (around 90%), but not all, are found in ExAC database (Supp. Table S2).
| Gene | FBN1 | TGFBR1 | TGFBR2 | SMAD3 | ACTA2 | MYH11 | MYLK |
|---|---|---|---|---|---|---|---|
| Number of variations reported (by events) | 484 | 66 | 72 | 76 | 30 | 407 | 387 |
| Exonic variations: number of variations | 245 | 31 | 49 | 34 | 21 | 270 | 255 |
| Missense | 140 | 12 | 28 | 13 | 1 | 141 | 147 |
| Stop gained | 0 | 0 | 1 | 0 | 0 | 0 | 2 |
| Frameshift insertion | 0 | 0 | 0 | 0 | 0 | 2 | 0 |
| Inframe insertion | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Frameshift deletion | 0 | 0 | 1 | 0 | 0 | 2 | 2 |
| Inframe deletion | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Synonymous | 105 | 19 | 19 | 21 | 20 | 125 | 104 |
| Intronic variations: number of variations | 237 | 34 | 21 | 35 | 9 | 136 | 132 |
| Intronic | 208 | 32 | 16 | 33 | 8 | 99 | 101 |
| ncRNA | 0 | 0 | 0 | 0 | 0 | 9 | 12 |
| Splice regions | 29 | 2 | 5 | 2 | 1 | 28 | 19 |
| 3' UTR | 2 | 1 | 2 | 3 | 0 | 0 | 0 |
| 5' UTR | 0 | 0 | 0 | 4 | 0 | 1 | 0 |
| Gene | FBN1 | TGFBR1 | TGFBR2 | SMAD3 | ACTA2 | MYH11 | MYLK | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | By events | Total | By events | Total | By events | Total | By events | Total | By events | Total | By events | Total | By events | |
| Number of variations | 364,879 | 2,365 | 40,976 | 360 | 92,387 | 464 | 34,076 | 384 | 23,880 | 229 | 733,666 | 1,896 | 1,065,693 | 1,708 |
| Exonic variations | 169,295 | 1,143 | 1,816 | 162 | 10,828 | 264 | 7,570 | 156 | 389 | 115 | 373,471 | 1,202 | 649,945 | 1,096 |
| Missense | 128,617 | 703 | 412 | 88 | 1,222 | 147 | 5, 133 | 61 | 101 | 54 | 40,018 | 676 | 301,731 | 681 |
| Stop gained | 10 | 4 | 1 | 1 | 3 | 3 | 2 | 2 | 0 | 0 | 8 | 8 | 23 | 17 |
| Frameshift insertion | 1 | 1 | 1 | 1 | 2 | 2 | 1 | 1 | 2 | 2 | 27 | 22 | 13 | 12 |
| Inframe insertion | 8 | 4 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 17 | 11 | 2 | 2 |
| Frameshift deletion | 18 | 18 | 4 | 4 | 9 | 9 | 6 | 6 | 6 | 6 | 23 | 23 | 30 | 28 |
| Inframe deletion | 0 | 0 | 3 | 3 | 4 | 3 | 2 | 2 | 0 | 0 | 31 | 11 | 8,363 | 17 |
| Synonymous | 40,641 | 413 | 1,395 | 65 | 9,587 | 99 | 2,425 | 83 | 280 | 53 | 333,347 | 451 | 339,783 | 339 |
| Intronic variations | 195,435 | 1,200 | 39,124 | 186 | 81,529 | 179 | 14,080 | 196 | 23,491 | 114 | 360,176 | 685 | 415,746 | 610 |
| Deep intronic | 169,628 | 1,076 | 39,112 | 177 | 944 | 162 | 14,047 | 182 | 23,478 | 107 | 355,875 | 553 | 324,260 | 490 |
| ncRNA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 418 | 32 | 29,999 | 33 |
| Splice regions | 25,807 | 124 | 12 | 9 | 80,585 | 17 | 33 | 14 | 13 | 7 | 3,883 | 100 | 61,487 | 87 |
| 3' UTR | 67 | 8 | 35 | 11 | 17 | 10 | 29 | 16 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5' UTR | 82 | 14 | 1 | 1 | 13 | 11 | 12,397 | 16 | 0 | 0 | 19 | 9 | 2 | 2 |
To predict mutations affecting splicing signals we applied the reference Human Splicing Finder (HSF)® system [Desmet et al., 2009]. Missense variations have been evaluated with UMD-Predictor [Frédéric et al., 2009; Salgado et al., 2016]. Nonsense mutations, out-of-frame insertions, or deletions were considered as pathogenic. Other variations (deep intronic, in 5’ and 3’ UTRs, in frame insertion and deletion) were not taken into account, as prediction tools are not sufficiently accurate and such mutations have only rarely been demonstrated to be pathogenic. As each identified variation is not linked to a specific sample (patient confidentiality), we were unable to approximate the number of variations per patient in each of these genes. In these seven genes, 2,222 different mutational events are predicted pathogenic and have been reported 10,014 times (Supp. Table S3).
When looking at ExAC-predicted pathogenic mutations not found in UMD databases, allele frequencies are lower than frequencies of ExAC-predicted pathogenic variations found in UMD databases (Supp. Fig. S1). These frequencies are also lower than frequencies of ExAC-predicted nonpathogenic variations that display a wide range of frequencies (Supp. Fig. S2). They mainly correspond to rare events. Predicted pathogenic mutations in ExAC found in UMD databases should represent only the most frequent events from UMD-databases and the numerous UMD databases pathogenic mutations not found in ExAC should correspond to very rare events not caught by random WES. Nevertheless, in the 67 FBN1 mutational events reported in ExAC and in the UMD-FBN1 database (Table 1), the most frequent FBN1 mutations are not found as c.5788+5G>A mutation reported in 30 nonrelated patients (30x), c.7754T>C (30x), c.247+1G>A (18x), c.1633C>T (17x), c.4588C>T (16x), c.8176C>T (16x), c. 364C>T (16x), c.7039_7040delAT (16x), or c.1879C>T (16x).
FBN1, TGFBR1 and 2, ACTA2, SMAD3, MYH11, MYLK, and Cardiovascular Risks
The identification of at risk patients is of major importance for the patient and their relatives for management, surveillance, as well as for genetic counselling purposes. Indeed, the natural history of asymptomatic ascending aortic aneurysms is progressive enlargement over time and ultimately life-threatening acute aortic dissection. With proper management, including medical therapy and prophylactic repair of an aneurysm, the life expectancy of an individual with a thoracic aortic aneurysm should approach that of the general population. This has already been observed with FBN1 and TGFBR2 gene mutations [Attias et al., 2009]. Therefore, the first interest for genetic testing is medical and surgical management of patients. Some recommendations are common to all entities, such as the prescription of medications that reduce hemodynamic stress on the aorta [Erbel et al., 2014], such as beta adrenergic blocking agents, but others can be modified according to the gene involved. Indeed, if the risk of aortic dissection increases at a maximal aortic dimension of about 5.5 cm in some presentations [Davies et al., 2002], aortic dissections have been reported in individuals with aortic diameters of less than 5.5 cm, or even 5.0 cm in others [Milewicz et al., 1998; Loeys et al., 2006; Zhu et al., 2006; Guo et al., 2007; Pannu et al., 2007; Pape et al., 2007; Tran-Fadulu et al., 2009]. In particular, early prophylactic repair should be discussed in individuals with an early-onset severe presentation and confirmed mutations in TGFBR2 and TGFBR1 and/or a family history of aortic dissection with minimal aortic enlargement. It has also been discussed for patients with ACTA2, MYLK, and MYH11 mutations (Table 6) [Hiratzka et al., 2010].
| Gene | Associated phenotype (MIM) | Possible extra-aortic clinical features | Specificities for follow-up (in addition to ascending aortic imaging and betablockades) |
|---|---|---|---|
| FBN1 | Marfan syndrome (#154700) | Ectopia lentis, skeletal, skin, and lung abnormalities | Ophthalmological and skeletal follow-up |
| MASS syndrome (#604308) | |||
| TGFBR2 | Loeys-Dietz syndrome type 2 (#610168) | Other arterial aneurysms (brain, iliac, abdominal aorta), craniofacial, and skeletal abnormalities | Imaging of the cerebral circulation, descending thoracic and abdominal aorta, and arterial branches originating from the aorta |
| Aortic surgery discussed before the aorta reaches 4.5 cm in early onset cases | |||
| TGFBR1 | Loeys-Dietz syndrome type 1 (#609192) | Other arterial aneurysms (brain, iliac, abdominal aorta), craniofacial, skeletal, and skin abnormalities | Imaging of the cerebral circulation, descending thoracic and abdominal aorta, and arterial branches originating from the aorta |
| Aortic surgery discussed before the aorta reaches 4.5 cm in early onset cases | |||
| ACTA2 | Familial thoracic aortic aneurysm type 6 (#611788)Multisystemic smooth muscle dysfunction syndrome (#613834) | Livedo reticularis, iris flocculi, early onset occlusive vascular diseases (including coronary artery disease and stroke, as well as Moyamoya-like cerebrovascular disease), periventricular white matter hyperintensities on MRI, pulmonary hypertension, hypotonic bladder, and malrotation and hypoperistalsis of the gut | Cerebrovascular imaging to assess for cerebrovascular disease and cardiac evaluation to assess for coronary artery diseaseSurgery when the diameter of the ascending aorta is between 4.5 and 5.0 cm |
| Moyamoya disease 5 (#614042) | |||
| SMAD3 | Aneurysms-osteoarthritis syndrome (#613795) | Early-onset osteoarthritis | Skeletal survey |
| Other arterial aneurysms (brain, iliac, abdominal aorta) | Imaging of the cerebral circulation, descending thoracic and abdominal aorta, and arterial branches originating from the aorta | ||
| MYH11 | Familial thoracic aortic aneurysm type 4 (#132900) | Patent ductus arteriosus | Surgery when the diameter of the ascending aorta is between 4.5 and 5.0 cm |
| MYLK | Familial thoracic aortic aneurysm type 7 (#613780) | None | Early aortic dissections with minimal or no dilatation |
The same assumption could be made for the second interest for genetic testing, i.e. follow-up of patients. Again, the list of circumstances that should be avoided is common to all genetic predispositions to aortic dissection because they are associated with increased stress on the aorta (uncontrolled hypertension, isometric exercise, bodybuilding/weight training exercises, and competitive sports that could lead to a significant increase in blood pressure), but systematic imaging for additional vascular disease is based on the gene that is mutated and/or family history. Indeed, extra-aortic imaging is recommended in patients with TGFBR1, TGFBR2, ACTA2, SMAD3, and TGFB2 mutations (Table 6) [Loeys et al., 2006; LeMaire et al., 2007; Tran-Fadulu et al., 2009; Milewicz et al., 2010].
The third interest for genetic testing is genetic counseling. All cases of TAAD with known molecular bases are inherited following an autosomal dominant inheritance with variable expression, and with or without decreased penetrance. The children of an affected parent have an up to 50% chance of inheriting the genetic predisposition to TAAD, and, if a carrier, the same risk of transmitting the disease. The identification of the disease-causing variant is of major importance to establish at risk individuals that would benefit from specific surveillance, and, on the contrary, to avoid unnecessary follow-up and undue sport limitation. Also, in some severe presentations, prenatal testing or preimplantation genetic diagnosis could be discussed case-by-case, and also necessitate the identification of the disease-causing mutation in the family.
Discussion
We are currently facing a technological breakthrough with high-throughput sequencing accessibility revolutionizing patient management not only in a diagnostic context but also in the way we can decipher pathophysiological mechanisms. These technologies enable the emergence of tremendous quantities of genomic data and a variety of databases but many questions still need to be answered: Which data are now available? Are they accessible? What is their quality and accuracy? What is their value in a clinical exome context? and How can we retrieve evidences for pathogenicity of mutations?
In this revolutionary age, one can ask about the legitimacy of LSDB. On one hand, they are mainly maintained by small organizations (compared to core databases) rising for some of them the problems of updates and sustainability (websites or databases were sometimes not updated after project closure). In addition, to reach community adhesion to these projects and ensure data sharing from specialized centers to the LSDB is often an issue, as centers do not have dedicated staff for this time-consuming activity because of lack of funding. On the other hand, LSDB strengths are their high quality and accuracy, as mutations are first collected from teams involved in diagnostic or research, highly specialized in the gene of interest, and then validated by database curators. Through this process, matching on reference gene sequences and accurate mutation naming are ensured (for example inadequate mutation names represent today around 5% of FBN1 mutation reports in scientific literature). These databases are also of substantial importance for practitioners and biologists who have to interpret gene variations. They allow access to extensive information for answering questions such as: what is the mutational spectrum of this gene (in order to adapt screening techniques)? Has this variation been previously characterized? What are the evidences for its pathogenicity? Which team can I contact to have supplementary data on this variation? Finally, with the Ghent 2 nosology, it is now essential to know if the variation has already been described for a patient presenting with cardiovascular symptoms as aortic dilation/dissection. Relying on LSDB knowledgebases, relevant information may be rapidly collected for data interpretation, more accurate results may be reported and costly time saved.
In the more general context of genomics, the releases of the "1000 Genomes" database, the NHLBI-GO Exome Sequencing Project (ESP) and the Exome Aggregation Consortium (ExAC) (including the previous two) give access to aggregated and harmonized exome sequencing data from a variety of large-scale sequencing projects (ExAC: 60,706 unrelated individuals sequenced as part of various disease-specific and population genetic studies) and therefore the added-value of LSDBs is raised especially in the clinical exome context. To address this question, we have searched for all the variations listed in ESP and ExAC and matching reference transcripts of MSARD genes. As variations are not linked to a specific sample for evident patient confidentiality, we were not able to approximate the number of variations in each of these genes by experiment and therefore were not able to address the question of how many candidate mutations in MSARD genes will need to be evaluated per clinical exome.
We therefore focused on the predicted pathogenicity of reported mutations using the UMD-Predictor [Salgado et al., 2016] and HSF [Desmet et al., 2009] systems in order to address two questions: “Are pathogenic mutations from LSDBs frequently found in large-scale sequencing projects?” and “Do large-scale sequencing projects contain pathogenic mutations not reported in LSDB?”. When looking at pathogenic mutations described in UMD-LSDBs and also reported in ExAC, matching is very low (4%, 88/2,210 mutations, Table 1). Among these 88 variations, 67 have frequencies below the threshold of 10−4 indicating that large-scale sequencing projects such as ExAC have only captured a limited set of pathogenic mutations related to MSARD because of the still limited number (60,706) of exomes. In fact, a simple calculation can be performed considering the number of mutational events and the frequency of the disease. If considering MSARD has a frequency (f) of 1/5,000 and that more than 2,000 pathogenic mutations (n) have been reported in these genes, then to capture a specific mutation, one will need to screen  individuals. Note that this value is underestimated as, first, pathogenic mutations have various frequencies and many are private and second, many MSARD diseases have a lower prevalence.
individuals. Note that this value is underestimated as, first, pathogenic mutations have various frequencies and many are private and second, many MSARD diseases have a lower prevalence.
On the other hand, predicted nonpathogenic mutations reported in ExAC displayed a wide range of frequencies (Supp. Fig. S2), many of them being rare with an allele frequency <10−4 [Lek et al., 2016]. These results suggest that the ExAC allele frequency could be considered as evidence to select candidate pathogenic mutations but that this evidence is today weak in the case of MSARD gene mutations.
In the context of international initiatives to promote data sharing such as the International Rare Disease Research Consortium (IRDiRC, http://www.irdirc.org), the Global Alliance for Genomics & Health (http://genomicsandhealth.org), the Human Variome Project (http://www.humanvariomeproject.org), and ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/), and taking into account the need for high-quality resources to help clinical exome interpretation, LSDBs are today highly needed. In fact, they have many advantages compared to large-scale initiatives as they contain exhaustive sets of pathogenic mutations obtained from manually curated data linked to evidences of pathogenicity. The large-scale sequencing resources could today be considered as added value to rapidly identify nonpathogenic variations based on a high allele frequency (>0.01) but only provide weak evidence to identify pathogenic mutations. This may change, as these resources will include more data. Nevertheless, this may take time, as they will need to include data from millions of individuals to eventually capture a significant portion of pathogenic mutations ensuring that they are not biased by targeted disease sequencing projects. Even so, the allele frequency evidence might still be weak as no phenotypic information is linked to reported variations.
In the era of "secondary findings" in clinical practice and more specifically for MSARD, UMD-LSDBs are today key resources to collect relevant information for data interpretation, report more accurate results and save time. They will benefit from data sharing from diagnostic laboratories and researchers as it is anticipated that the amount of mutations identified by WES and WGS from these genes will explode in the coming years. UMD-LSDBs of MSARD genes could be viewed as beacons in the dark thanks to the high quality of data. This has already been recognized by other networks and resulted in private–public partnerships to promote and develop such resources as exemplified by the BRCAShare™ initiative (http://www.umd.be/BRCA1/) for UMD-BRCA1/2 in the context of breast cancers (Béroud et al. 2016). The BRCA1/2 genes also belong to the ACMG list of 56 actionable genes for which it is recommended to report findings to patients in a clinical exome context.
Acknowledgments
A.P. is supported by a PhD studentship from AFSMA (Association Française du Syndrome de Marfan et Apparentés).
Disclosure statement: The authors have no conflict of interest to declare.




