BRCA Share: A Collection of Clinical BRCA Gene Variants
For the Next Generation Sequencing special issue
ABSTRACT
As next-generation sequencing increases access to human genetic variation, the challenge of determining clinical significance of variants becomes ever more acute. Germline variants in the BRCA1 and BRCA2 genes can confer substantial lifetime risk of breast and ovarian cancer. Assessment of variant pathogenicity is a vital part of clinical genetic testing for these genes. A database of clinical observations of BRCA variants is a critical resource in that process. This article describes BRCA Share™, a database created by a unique international alliance of academic centers and commercial testing laboratories. By integrating the content of the Universal Mutation Database generated by the French Unicancer Genetic Group with the testing results of two large commercial laboratories, Quest Diagnostics and Laboratory Corporation of America (LabCorp), BRCA Share™ has assembled one of the largest publicly accessible collections of BRCA variants currently available. Although access is available to academic researchers without charge, commercial participants in the project are required to pay a support fee and contribute their data. The fees fund the ongoing curation effort, as well as planned experiments to functionally characterize variants of uncertain significance. BRCA Share™ databases can therefore be considered as models of successful data sharing between private companies and the academic world.
Introduction
Breast cancer is the most common malignancy in women in most countries with 464,000 new cases and 131,000 deaths each year in Europe and 246,660 new cases and 40,450 deaths in the US [Ferlay et al., 2013]. Globally, it is estimated that more than 1 million women worldwide are diagnosed yearly with breast cancer and that more than 400,000 will die from the disease. Although most cases are sporadic, familial clustering has been reported and it is estimated that approximately 10% are likely to be hereditary. The known pathogenic mutations** from predisposing genes, including BRCA1 (MIM# 113705) and BRCA2 (MIM# 600185), account for 20%–25% of those familial forms [Pharoah et al., 2002].
As breast cancers represent a significant public health problem, many national programs have been dedicated to cancer prevention and early diagnosis. They use various risk assessment (Gail [Chay et al., 2012], Claus [Fischer et al., 2013] and Tyrer-Cuzick [Boughey et al., 2010]) or probability models such as the BReast CAncer risk PRediction mOdel (BRCAPRO) [Mazzola et al., 2015]. The average cumulative risks in BRCA1 mutation carriers by the age of 70 are 65% (95% confidence interval 44%–78%) for breast cancer and 39% (18%–54%) for ovarian cancer. The corresponding estimates for BRCA2 mutation carriers are 45% (31%–56%) and 11% (2.4%–19%) [Antoniou et al., 2003]. Women with a high lifetime risk (≥20%) are eligible for genetic counseling and testing mainly for mutations of the BRCA1 [Hall et al., 1990] and BRCA2 [Wooster et al., 1994] cancer susceptibility genes.
Professional organizations in North America and Europe have published clinical practice guidelines with respect to BRCA counseling and testing [Balmaña et al., 2011; Gadzicki et al., 2011; Graña et al., 2011; National Institute for Health Care Excellence 2013; Cancer Institute NSW 2015; Holter et al., 2015]. The United States National Comprehensive Cancer Network guidelines recommend genetic counseling and/or testing for women that fulfill one of the following criteria: breast cancer at the age of 50 years or younger; a bilateral breast cancer; triple-negative breast cancer (estrogen receptor negative, progesterone receptor negative, Her2Neu negative); breast cancer at any age with close relatives with breast/ovarian/pancreatic cancer; breast cancer with Ashkenazi Jewish ancestry; male breast cancer; known mutation for a breast cancer susceptibility gene within the family, or women with a history of ovarian cancer. These recommendations have resulted in a rapid uptake and utilization of BRCA testing in clinical practice and a consequent large increase in variant classification burdens among laboratories worldwide.
The process of assessing clinical significance of germline variants has been codified in guidelines [Bahcall 2015] and formalized in probabilistic models for evidence integration [Goldgar et al., 2004; Lovelock et al., 2007]. Evidence may come from family studies, functional studies, algorithmic predictions, patterns of allelic co-occurrences within individuals, and other sources. Clinical significance is typically assessed, per the ACMG/AMP guidelines [Richards et al., 2015], by assigning the variant to one of the following classifications: Pathogenic, Likely Pathogenic, VUS (Variant of Uncertain Significance), Likely Benign, and Benign. Integrating evidence to produce a variant classification currently relies on the judgment of geneticists with diverse areas of expertise who primarily assess the information available from the scientific literature and databases, as well as algorithmic predictions, to arrive at a variant classification. Several studies reporting inter reviewer agreements in classifications among experts with widely differing outcomes ranging from “moderate” (Gross k, 0.52) to “high” (K-alpha, 0.91) have been published. Specifically, although a higher concordance of potentially clinically actionable classifications (Pathogenic or Likely Pathogenic) derived using the ACMG-AMP schemes with those reported in locus-specific databases (LSDBs) and ClinVar have been reported, the concordance rates for classifications deemed as non-actionable (VUS, Likely Benign, and Benign) were lower [Maxwell et al., 2016]. Amendola et al. (2016) reported a 79% concordance when nine participating laboratories classified a set of variants using their internally developed methods and the ACMG-AMP criteria. However, only a 34% concordance for either classification system across laboratories was observed. After consensus discussions and detailed review of the ACMG/AMP criteria, concordance increased to 71%, showing that a common framework can help resolve differences in classifications.
It can require many hours to classify a variant when literature review is required. The results and conclusions often need to be fully vetted with a critical eye toward translation to the mechanism(s) and presentation(s) of the disease and phenotype under consideration. Understandably, this classification adds significant skilled labor cost to the testing process. Fully automated methods for variant classification are not ready for clinical practice. Median time required to curate available lines of evidence and classify variants have been reported to vary between 54 min (range 5–233 min) [Dewey et al., 2014], to 37 min per variant (range 1–175 min) [Amendola et al., 2015]. Furthermore, variant classifications are not static; classifications may change over time as the underlying scientific knowledge changes, requiring a periodic re-evaluation of the literature and other evidence prior to clinical reporting. CLIA laboratory regulations in the United States require each laboratory to be responsible for its own reports, and to update their classifications to reflect the most recent information, which limits the ability to use a paradigm for static classifications (and hence share the cost of classification) across laboratories.
Variant databases play a critical role in the classification process, by providing summary information about the state of scientific knowledge, with clinical assessments of the variant by other laboratories, with supporting evidence, and with data on other patients carrying the variant. LSDBs [Claustres et al., 2002] have been established for many disease genes, whereas other databases, including OMIM, ClinVar, LOVD, and Clinvitae collect variant information across genes. Recently, network federation protocols such as Beacon [Krol 2015] and Matchmaker exchange [Philippakis et al., 2015] have been established to facilitate peer-to-peer exchange of variant data. Some commercial software products, including GeneInsight Network and Agilent's Cartagenia, are using a similar approach to enable sharing of variant classifications within their user communities.
In the context of BRCA testing, databases available today include: the Breast Cancer Information Core Database (BIC) [Szabo et al., 2000], LOVD [Fokkema et al., 2011], ClinVar [Landrum et al., 2014], BRCA Exchange [Global Alliance for Genomics and Health 2016], the ARUP databases (www.arup.utah.edu/), and the Universal Mutation Database (UMD)-BRCA1/2 databases [Caputo et al., 2012]. Note that Myriad Genetics, a commercial laboratory, has developed its own database whose quality and content could not be evaluated as it is proprietary and not accessible to the community. These databases have been developed with different goals and have different curation processes, data quality, and content as underlined by Vail et al. [2015]. These authors compared BIC, ClinVar, HGMD (paid version), LOVD, and the UMD databases and concluded that these differences inhibit their wider use in clinical practice.
In order to provide high-quality sustainable BRCA1 and BRCA2 databases, the BRCA Share™ public/private partnership was co-founded by Quest Diagnostics and the French National Institute of Health and Medical Research (Inserm) in April 2015, with Laboratory Corporation of America Holdings as the first commercial participant. The goal of the initiative is to share clinical, genetic, epidemiological, and biological data on BRCA variants, particularly VUS, in order to improve the quality of laboratory diagnostics to better predict which individuals are at risk of developing hereditary breast and ovarian cancers, and to accelerate research on BRCA gene variants. BRCA Share™ builds on an efficient data curation process [Caputo et al., 2012] that follows international recommendations [Eccles et al., 2015].
The BRCA Share™ database now contains over 6,200 total BRCA variants, an increase of nearly 30% compared with the previous UMD-BRCA1/2 databases. Of these variants, 334 are newly identified pathogenic or likely pathogenic, increasing by about 20% the total number of pathogenic or likely pathogenic variants to 1,826.
To our knowledge, BRCA Share™ is the first example of a successful public/private partnership for LSDBs that ensures free access to the database for research purposes while creating a user group of commercial partners that will collectively endorse running and development costs while also supporting functional tests to rapidly and efficiently classify VUS.
With increased adoption of whole exome and whole genome sequencing in clinical practice, it is believed that many patients might benefit from the indirect discovery of variants in clinically actionable genes. In a study of 1,000 individuals (500 European- and 500 African-descent participants randomly selected from the National Heart, Lung, and Blood Institute Exome Sequencing Project), Dorschner et al. [2013] report that ∼3.4% of European-descent adults and ∼1.2% of African-descent adults can be expected to have actionable highly penetrant pathogenic or likely pathogenic mutations identified by exome sequencing. Among those, three of the 1,000 participants had a BRCA1/2 pathogenic mutation [Dorschner et al., 2013]. This ratio was confirmed by a subsequent study by Amendola et al. [2015].
With the involvement of the major diagnostic companies in US and reference centers in France, for the first time, all variants will be shared and further annotated by a group of experts [Caputo et al., 2012]. The availability of this database is expected to provide researchers and geneticists rapid access to supporting evidence for classification.
Materials and Methods
The BRCA Share Databases
The BRCA Share™ databases are derived from the UMD-BRCA1 and UMD-BRCA2 databases [Caputo et al., 2012], based on 20 years’ experience of clinical BRCA testing by UGG, and augmented by the results of clinical BRCA testing by Quest and LabCorp. The UMD software [Béroud et al., 2005] has been modified to accommodate new features including a registration process and secure access to the databases under individual login IDs.
In order to integrate data from different sources and ensure an optimal curation process, a common template was used for data submission. It contains information related to the individual and related sample(s), including submitter, de-identified subject and family IDs, subject demographic information, disease status of subject and relatives, availability of cell line, tumor, or other physical materials, and so on. Subject and family identifiers are reassigned to yield globally unique anonymous identifiers which can be mapped back to the submitter's anonymous identifiers for future updates. Information related to variants includes the next-generation sequencing (NGS) screening type (whole coding sequence; targeted; single site), sequencing platform, DNA, RNA and protein HGVS names for the variant [den Dunnen et al., 2016], ID of transcript used to derive the names, the variant class (missense, nonsense, frameshift, intronic, rearrangement, insertion, deletion, isosemantic a.k.a. synonymous), the submitter pathogenicity assessment, and supporting evidence collected from the literature, from functional assays and from in silico tools. Variant nomenclature is automatically validated by the UMD software and the HGVS genomic DNA variant name (g.) is generated using the GRCh37 reference sequence. Allele frequencies are automatically collected from dbSNP [Sherry et al., 2001], the Exome Aggregation Consortium including TCGA data (http://exac.broadinstitute.org/), the 1000 genomes and the 6,500 exomes of the Exome Variant Server (http://evs.gs.washington.edu/EVS/). Finally, a link to the UMD-Predictor system [Salgado et al., 2016] is provided to ensure access to the most recent in silico predictions from this system.
The BRCA Share™ databases are accessible at http://umd.be/BRCA1/ and http://umd.be/BRCA2/. They integrate a new Web interface that uses dynamic tabs and forms. It was developed using the JQuery library (http://jquery.com/). New graphical displays were also created, using the D3.js library (http://d3js.org/), featuring dynamic zoom and signals highlights.
As an example of the depth of annotation provided by these databases, consider the BRCA2 c.316+5G>C (IVS3+5G>C) variant, listed as pathogenic in BRCA Share™. Figure 1 shows a snippet from BRCA Share™ for this mutation. It provides brief summaries for descriptions in the published literature, in silico splice predictions, frequency, co-occurrence, and even classifications from other public databases. Supporting evidence for pathogenic classification including references to functional studies describing aberrant splicing, frequency, and co-occurrence in patients are included in the variant record. This allows the investigator to review current data and literature for validation of the variant's classification and not solely rely on the classification from other clinical laboratories.
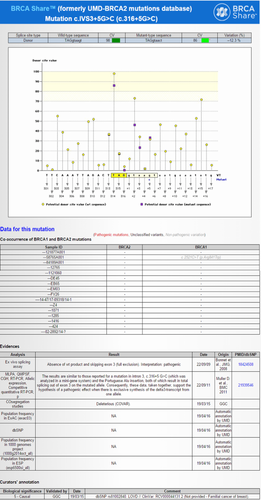
Two new features have been added on the welcome page to highlight changes in classification status: the "Variants reclassification" and "Variants classification" links. The first one gives access to the list of reclassified variants during the last 6 months, whereas the second gives access to variants that have been classified during the last 6 months. Note that during the registration process, the user can select to receive alerts by email when variant reclassification occurs.
Curation
BRCA Share curation is based on processes developed by the Unicancer Genetic Group (UGG) BRCA network that ensure clinical grade quality. It relies on a classification working group which meets regularly to provide expert curation. VUS classification is conducted using a combination of available data: frequencies in the general population, in silico predictions using bioinformatics tools at both splice and protein levels, causality or neutrality scores published in the literature [Goldgar et al., 2004; Easton et al., 2007], functional domain [Millot et al., 2012; Guidugli et al., 2013], co-occurrence with causal mutations in the same gene, co-segregation analyses in French families and published results of functional tests or splicing modeling data as described in Caputo et al. [2012]. Evidence for classification are integrated in a likelihood model previously described [Goldgar et al., 2004]. The classifications displayed in the database are limited to those derived by the UGG. Periodically, discordant classifications among participating laboratories are marked with an asterisk to alert the user toward a closer review.
Results
Usage and Content Statistics
The BRCA Share databases have been accessible online since July 2015. In this first year, the number of registrants has grown regularly to reach 1,119 users and the sites have been queried on average 17,000 times/month.
As a result of the establishment of the BRCA Share™ effort and the addition of clinical variants from Quest and LabCorp, the number of BRCA1 and BRCA2 variants has grown from UMD's total of 4,838 at the time BRCA Share was launched, to its current total of 6,254 unique variants, an increase of 29.3%.
The current count of records and variants are shown in Tables 1 and 2, and broken down by pathogenicity assessment and variant type, respectively. The distribution by variation type and pathogenicity assessment is available in Supp. Table S1.
| BRCA1 | BRCA2 | Combined | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Records | Unique variants | Records | Unique variants | Records | Unique Variants | |||||||
| 1. Neutral (Benign) | 70,388 | 88.2% | 114 | 4.5% | 45,221 | 78.3% | 123 | 3.3% | 115,609 | 90.2% | 237 | 6.4% |
| 2. Likely Neutral (Likely Benign) | 944 | 1.2% | 142 | 5.6% | 2,487 | 4.3% | 218 | 5.9% | 2,487 | 1.9% | 360 | 9.7% |
| 3. VUS | 2,828 | 3.5% | 1,423 | 56.0% | 5,742 | 9.9% | 2,408 | 64.9% | 5,742 | 4.5% | 3,831 | 103.2% |
| 4. Likely causal (Likely Pathogenic) | 39 | 0.0% | 21 | 0.8% | 46 | 0.1% | 34 | 0.9% | 46 | 0.0% | 55 | 1.5% |
| 5. Causal (Pathogenic) | 5,626 | 7.0% | 843 | 33.1% | 4,276 | 7.4% | 928 | 25.0% | 4,276 | 3.3% | 1,771 | 47.7% |
| Total | 79,825 | 2,543 | 57,772 | 3,711 | 128,160 | 6,254 | ||||||
| BRCA1 | BRCA2 | Combined | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Records | Unique mutations | Records | Unique mutations | Records | Unique mutations | |||||||
| # | % | # | % | # | % | # | % | # | % | # | % | |
| Total | 79,825 | 2,543 | 57,772 | 3,711 | 137,597 | 6,254 | ||||||
| Large rearrangements (>1 exon) | 458 | 0.6% | 79 | 3.1% | 59 | 0.1% | 32 | 0.9% | 517 | 0.4% | 111 | 1.8% |
| Deletions | 372 | 0.5% | 58 | 2.3% | 37 | 0.1% | 24 | 0.6% | 409 | 0.3% | 82 | 1.3% |
| Insertions | 86 | 0.1% | 21 | 0.8% | 22 | 0.0% | 8 | 0.2% | 108 | 0.1% | 29 | 0.5% |
| Small deletions and insertions | 3,145 | 3.9% | 531 | 20.9% | 3,115 | 5.4% | 702 | 18.9% | 6,260 | 4.5% | 1,233 | 19.7% |
| Small deletions | 2,234 | 2.8% | 389 | 15.3% | 2,538 | 4.4% | 530 | 14.3% | 4,772 | 3.5% | 919 | 14.7% |
| Small insertions | 911 | 1.1% | 142 | 5.6% | 577 | 1.0% | 172 | 4.6% | 1,488 | 1.1% | 314 | 5.0% |
| Point mutations | 73,420 | 92.0% | 1,264 | 49.7% | 38,395 | 66.5% | 2,195 | 59.1% | 111,815 | 81.3% | 3,459 | 55.3% |
| Missense | 72,227 | 90.5% | 1,068 | 42.0% | 37,185 | 64.4% | 1,964 | 52.9% | 109,412 | 79.5% | 3,032 | 48.5% |
| Nonsense | 1,193 | 1.5% | 196 | 7.7% | 1,210 | 2.1% | 231 | 6.2% | 2,403 | 1.7% | 427 | 6.8% |
| Intronic mutations | 2,593 | 3.2% | 628 | 24.7% | 16,103 | 27.9% | 743 | 20.0% | 18,696 | 13.6% | 1,371 | 21.9% |
| Splice sites (<10 bp from exon) | 809 | 1.0% | 167 | 6.6% | 827 | 1.4% | 172 | 4.6% | 1,636 | 1.2% | 339 | 5.4% |
| Mid-intronic mutations | 1,784 | 2.2% | 461 | 18.1% | 15,276 | 26.4% | 571 | 15.4% | 17,060 | 12.4% | 1,032 | 16.5% |
| Indels | 209 | 0.3% | 41 | 1.6% | 100 | 0.2% | 39 | 1.1% | 309 | 0.2% | 80 | 1.3% |
Overlap and Classification Agreement with other Collections
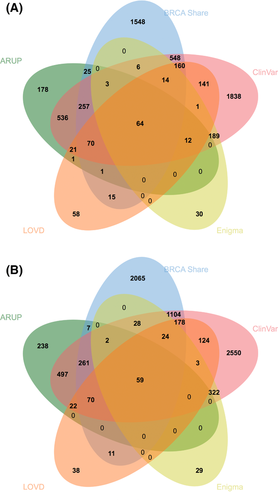
The Venn diagrams in Figure 2(A and B) show the counts of BRCA1 and BRCA2, variants, respectively, shared with ARUP (www.arup.utah.edu/), ClinVar [Landrum et al.,2014], the ENIGMA collection [Spurdle et al., 2012], and LOVD [Fokkema et al., 2011]. The BRCA Share™ collection currently is the second largest, behind ClinVar; both contain a large number of variants not present in the other. Note that ARUP only reports class 5 variants; its overlap with other systems would be more favorable if limited to these. Discordant classifications (2 or more classes difference) within ClinVar were treated as VUS for the purposes of comparison. Classification comparisons between the two largest sites, BRCA Share™ and ClinVar, are shown in Figure 3. Seventy-four percent of the variants are classified identically between these two sites. None of the discordances are of the clinically actionable type where a variant goes from likely pathogenic/pathogenic to likely benign/benign. Total VUS discrepancies are shown in the margins. The largest discordance categories are for variants classified as class 2 (Likely Benign) in ClinVar versus class 3 (VUS) in BRCA Share™ and class 5 (Pathogenic) in BRCA Share™ versus class 3 (VUS) in ClinVar. In addition, the comparison of variants classified as class 4/5 in BRCAShare™, ARUP, and ClinVar reveals that BRCAShare™ and ARUP are more in agreement with each other (499/546; 91.4%) than ARUP and ClinVar (400/1,062; 37.7%), possibly reflecting small differences in classification criteria.
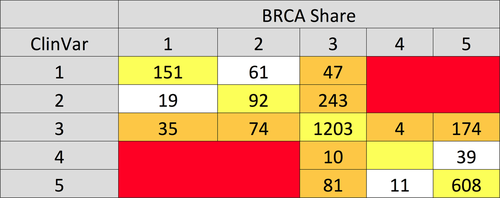
Pairwise comparisons among BRCA Share™, ClinVar, and ARUP show that BRCA Share™ and ClinVar agree on 72% of classifications, BRCA Share™ and ARUP on 81%, and ARUP and ClinVar on 60% of shared variants. Twenty four percent of variants classified as VUS by BRCA Share™ are classified as some other category by ClinVar, whereas 19% of variants classified as VUS by ClinVar are classified otherwise by BRCA Share™.
Variant Frequencies
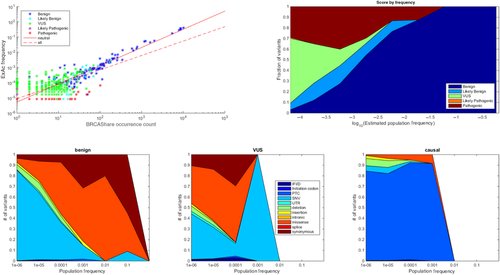
The BRCA Share™ database affords an opportunity to characterize the spectrum of BRCA variation in some detail, to gain understanding the relationships between frequency, type, and classification. Although population frequency is not directly available from the BRCA Share™ collection, it can be estimated from the database. This collection incorporates a number of biases of sampling and reporting, including emphasis on affected subjects and their relatives, leading to an emphasis of deleterious alleles. We estimated population frequencies for BRCA Share™ variants using a log–log regression of frequencies from the Exome Aggregation Consortium (ExAC) dataset against BRCA Share™ occurrence counts. The ExAC frequencies are based on a collection of 60,706 unrelated individuals sequenced as part of various disease-specific and population genetic studies for which an aggregate frequency is provided [http://exac.broadinstitute.org/]. As ExAC is not expected to be enriched for obligate carriers from families with a strong history of breast and ovarian cancer, the contribution of pathogenicity biases within this dataset are minimized. As shown in Figure 4(A), BRCA Share™ occurrence counts correlate well with aggregated ExAC frequencies overall, especially for common variants. The distribution of pathogenic variants is skewed higher in BRCA Share™ compared with ExAC, presumably reflecting the collection biases. A regression on benign variants only provides better agreement over most of the dynamic range than based on all variants. This regression was used to estimate frequencies for all variants, including those not shared with ExAC, in Figure 4(B–D). Figure 4B shows a breakdown of classifications in log10 estimated frequency bins, showing the increase in VUS and pathogenic rates among rare variants. Note that sample size effects cause the left most bin to be a truncation of unobserved rarer frequency bins to the left. Figure 4(C) shows the fractional representation of variant types in each log frequency bin, whereas Figure 4(D) shows the same for VUS only.
Co-occurrences
Co-occurrence of a VUS with a known disease-causing mutation, either from the same or the other BRCA gene, can provide evidence to classify the variant, especially if it is reported in multiple patients [Goldgar et al., 2008; Cherbal et al., 2012; Santos et al., 2014]. Co-occurrence data are used in two ways: in a Posterior Probability calculation [Goldgar et al., 2004], as it is one of its elements, and as a standalone criterion for classification. Indeed, if a given variant co-occurs with at least two different pathogenic variants in the same gene or is demonstrated co-occurring in trans with a pathogenic variant of the same gene, it is classified as Neutral. This criterion requires that the patient has been examined by a clinical geneticist to exclude Fanconi-like for BRCA2.
Since the BRCA Share™ databases preserve patient-centric information, they allow co-occurrences to be readily queried, as shown in Figure 5. During curation, such evidence can provide arguments against pathogenicity, as illustrated in Table 3. For each BRCA1 VUS from a specific sample, the table displays co-occurring pathogenic or likely pathogenic BRCA1 mutations. We can distinguish cases in which the same pair of variants co-occur repeatedly, as illustrated for BRCA2 c.324T>C (p.Asn108Asn), which is specifically associated with the c.2612C>A (p.Ser871X) pathogenic mutation. This might suggest a common origin of the patients or the presence of both variants on the same allele. By contrast, the BRCA2 c.2350A>G (p.Met784Val) variant was reported in two patients from different laboratories. Each of them also harbors a different pathogenic BRCA2 mutation: c.6952C>T (p.Arg2318X) and c.5771_5774delTTCA (p.Ile1924ArgfsX38). This indicates that those mutations are probably in trans and that these patients are unrelated. Additionally, this is a strong evidence to consider the BRCA2 c.2350A>G (p.Met784Val) variant as non-pathogenic. Thus, the co-occurrence data facilitate weighting of evidence based upon the number of samples displaying a co-occurrence with another pathogenic variant.
| HGVS c. | HGVS p. | Sample ID | Class | Pathogenic BRCA1 mutation |
|---|---|---|---|---|
| c.28A>G | p.Thr10Ala | 04-3625 | 3 | c.2701delC (p.Ala902LeufsX2) |
| c.324T>C | p.Asn108Asn | 02-2012 | 3 | c.2612C>A (p.Ser871X) |
| c.324T>C | p.Asn108Asn | 31-3504 | 3 | c.2612C>A (p.Ser871X) |
| c.324T>C | p.Asn108Asn | 01-65884A001 | 3 | c.2612C>A (p.Ser871X) |
| c.324T>C | p.Asn108Asn | 14-829 | 3 | c.2612C>A (p.Ser871X) |
| c.2320A>G | p.Thr774Ala | 15-676 | 3 | c.5576_5579delTTAA (p.Ile1859LysfsX3) |
| c.2350A>G | p.Met784Val | 01-82103A001 | 3 | c.6952C>T (p.Arg2318X) |
| c.2350A>G | p.Met784Val | 02-20417 | 3 | c.5771_5774delTTCA (p.Ile1924ArgfsX38) |
| c.2416G>C | p.Asp806His | FYxqoGbWhaFkaYn | 3 | c.IVS6-2A>G (c.476-2A>G) |
| c.2751A>G | p.Val917Val | 20-14YW68IE | 3 | c.8904delC (p.Val2969CysfsX7) |
| c.2780T>C | p.Met927Thr | 07-A633 | 3 | c.1310_1313delAAGA (p.Lys437IlefsX22) |
| c.2837A>G | p.Asp946Gly | 33-008FCF1006/33-4-292 | 3 | c.2701delC (p.Ala902LeufsX2) |
| c.2919G>A | p.Ser973Ser | 12-FK16 | 3 | c.1184G>A (p.Trp395X) |
| c.3152T>C | p.Leu1051Ser | 01-62951A001 | 3 | c.6656C>G (p.Ser2219X) |
| c.3170A>G | p.Lys1057Arg | 05-026081 | 3 | c.IVS15+1G>A (c.7617+1G>A) |
| c.3226G>A | p.Val1076Ile | 33-1-5524 | 3 | c.3159delA (p.Asp1054IlefsX6) |
| c.3304A>T | p.Asn1102Tyr | pfayNyctYdTDDjM | 3 | c.1929delG (p.Arg645GlufsX15) |
| c.3304A>T | p.Asn1102Tyr | VrnAyEGaCFDesIG | 3 | c.1929delG (p.Arg645GlufsX15) |
| c.3445A>G | p.Met1149Val | 11-2009-012 | 3 | c.2092delC (p.Leu698TyrfsX32) |
| c.3539A>G | p.Lys1180Arg | 02-11705 | 3 | c.8070_8071dup (p.Ser2691PhefsX4) |
- a First column: cDNA HGVS nomenclature of the mutation; second column: protein HGVS nomenclature of the mutation; third column: unique sample ID; fourth column: mutation class (3 = VUS); fifth column: HGVS c. and p. nomenclature of BRCA1 pathogenic mutations co-occurring in each sample.
Classification Discordances
The initial combined dataset contained 687 variants that were shared between two or more of the three participating laboratories, 67% (N = 457) of which were in BRCA1 and 33% (N = 230) were in BRCA2. Classifications were identical across the laboratories for 57% of the shared variants, whereas 69% were concordant when Likely Benign is grouped with Benign and Likely Pathogenic with Pathogenic. The remaining 31% included variants that crossed over from a VUS to a non-VUS category between laboratories. No drastic differences in classifications (i.e., Benign/Likely Benign vs. Pathogenic/Likely Pathogenic) with a potential to adversely impact patient outcomes were identified. Concordance rates were higher in BRCA1 than BRCA2, for example, exact concordances were 76% and 53%, respectively. We emphasize that these are per variant discordance rates and not per patient; since the discordances involve rare variants, per patient discordance rates would be far lower.
A process to resolve these discordances is underway, involving periodic review of the classification evidence by all the contributing laboratories. Initial review of 148 BRCA1 discordances revealed that most were attributable to changes in the available evidence since the original classification; after review the number was significantly reduced to 37, or 8.1% of the BRCA1 shared variants. Of these 37 discordances, 17 were attributable to differences in the approach toward classification of rare synonymous variants in the absence of other supportive evidence, with some groups classifying these as VUS and others as Likely Benign. The remaining 20 variants (4.4%) are all missense (18) or intronic (2), and are still under review, as are the BRCA2 variants. BRCA Share displays a primary classification for each variant, though submitter classifications are accessible. The primary classification displayed is from the French Consortium (UGG) if available or else the closest to VUS of the submitted variants is displayed. Discordant classifications among participating laboratories are marked with an asterisk.
Discussion
Since its inception a little over a year ago, the BRCA Share™ initiative has significantly augmented the UMD dataset collected by the UGG, by leveraging the clinical sample flow of two large laboratories. The combination of US and European clinical samples has likely increased the ethnic diversity of the BRCA Share™ sample, while still leaving large portions of human diversity undersampled.
The fact that BRCA Share™ and ClinVar, the two largest collection sites, each have a substantial number of variants not present in the other, indicates that no existing collection has yet to uncover all the variants in the human population.
Classification agreement within the participating BRCA Share™ groups was generally higher than previous reports, elsewhere, with most discordances being attributable either to incorporation of newer data or likely versus VUS differences.
The BRCA Share™ funding strategy, in which academic researchers have free access, while commercial labs pay support fees, may provide a sustainable model in a time when public funding for biological databases is under stress [Check Hayden 2016; Kaiser 2016]. Public collections such as ClinVar and GA4GH provide an alternative model with fewer restrictions on access, but they rely on government funding. ClinVar has historically functioned as a submission archive, without requiring resolution of conflicting pathogenicity classifications between different submitters, instead showing all submitted classifications for each variant. Submitters are encouraged to provide classification evidence but are not required to. Consequently, the quality of data curation can vary significantly from one variant to the next. ClinVar has implemented a star rating system to provide an indication of annotation quality. The NIH-funded ClinGen curation effort, in collaboration with ClinVar [https://www.clinicalgenome.org/data-sharing/clinvar/] is encouraging the formation of expert panels to resolve discordances with the intention to create over time a higher quality subset of ClinVar content that will be more readily usable in the clinic.
GA4GH [Global Alliance for Genomics and Health 2016] has recently announced the creation of BRCAExchange [http://brcaexchange.org/], a consolidated database that integrates all publicly available datasets on BRCA gene variants. The quality assurance and funding models for BRCAExchange are still under development. BRCA Share, by contrast, builds on the curation model developed by the French National Working Group on BRCA VUS Classification, and funds curation efforts from membership dues. As with ClinVar, complete sharing of BRCA Share™ data with BRCA Exchange would undermine the BRCA Share™ model, but sharing of variant content, either directly or via the distributed Beacon [Krol 2015] mechanism, is under discussion.
A portion of the BRCA Share™ funding is earmarked for functional studies to reduce the fraction of BRCA classifications in the variants of uncertain significance category. BRCA functional assays include assessments of E3-ligase activity, BARD1 binding and homology-directed repair [Starita et al., 2015], splicing [Théry et al., 2011], gene expression [Findlay et al., 2014], nuclear localization and P53 phosphorylation in response to DNA damage [Loke et al., 2015], growth restoration in BRCA1-deficient mouse embryonic stem cells [Bouwman et al., 2013], and others. Although there is not yet a consensus as to which of these will best correlate with in vivo phenotypes, the rapid recent progress provides hope that such a consensus may soon be forthcoming.
- Per patient, in which a functional study is performed as part of or as a follow up to the BRCA test of a particular patient specimen with the intent of improving the report for that patient [e.g., Loke et al. 2015]. This model is more appropriate for clinical labs than for a collection.
- Prioritized list, in which variants in a database are prioritized for functional studies according to some cost/benefit criteria.
- Exhaustive, in which all possible variants are generated via high-throughput mutagenesis and characterized in a high-throughput assay [Bouwman et al. 2013; Guidugli et al. 2013; Findlay et al. 2014; Starita et al. 2015; Sun et al. 2016].
An additional dimension for the design of functional studies is whether they require live patient cells or extracted DNA, or whether an in silico description of the variant will suffice. The high-throughput methods tend to require only in silico descriptions. Prioritized lists are compatible with both approaches but must be coupled to a cell, tissue or DNA bank, such as kConfab (http://www.kconfab.org/). No such collection currently exists for BRCA Share™.
We are piloting the prioritized list model using BRCA Share™ data. One obvious prioritization criterion is frequency; given a fixed capacity for doing functional studies, choosing more frequent variants will impact more patients. Since most VUS are rare, the impact of a functional study for a single variant on the total per patient VUS rate will necessarily be minimal; many VUS must be classified to have an impact. Moreover, under current ACMG Guidelines [Richards et al., 2015] functional studies can provide “strong” but not “very strong” evidence, which is insufficient to reclassify a variant as likely pathogenic or benign without 1–2 additional pieces of moderate strength evidence, or >2 pieces of supporting evidence. Probabilistic approaches to evidence integration may provide stronger weighting to functional studies based on demonstrated predictive value, but there is not yet a consensus on how to do this [Grandval et al., 2013].
The BRCA Share™ team has developed a prioritized list available from the "Statistics" section of the Website using the "Prioritization of VUS for validation" feature. It combines frequency (number of cases), available evidence, and co-occurrence data to produce a prioritization score, a high value means that additional evidence is available and that the addition of a functional study is likely to bring about reclassification by ACMG guidelines. In the future this may be used to prioritize allocation of BRCA Share™ funding for functional studies. Funding of high-throughput studies or construction of reagents to support them is also under consideration.
Such efforts will ultimately provide high-quality databases for actionable genes for which patients might directly benefit from adequate variant annotation during high-throughput sequencing analysis especially in the context of WES and WGS. We believe that such databases will still be needed in the future as they provide accurate annotations as well as the list of available evidence to help the end-user interpret difficult variants. They will exist in parallel to general databases such as ExAC dedicated to variant frequency. BRCA1/2 BRCA Share™ databases could today be considered as models to demonstrate that data sharing is now a reality between private companies and the academic world; that resulting data are shared with research teams as well as commercial partners engaged in the long-term sustainability of such initiatives; that strong efforts are dedicated to reclassify VUS; and finally that such systems might facilitate NGS handling and secondary findings interpretation.
Finally, although BRCA Share™ is currently limited to BRCA1 and 2, there are other clinically important genes that could benefit from a similar alliance between researchers and commercial labs. A useful next step would be to include other important cancer risk genes into a Cancer Risk Share.
Disclosure statement: The authors declare no conflict of interest.




