Journal list menu
Export Citations
Download PDFs
The TP53 Gene Network in a Postgenomic Era
- Pages: 641-642
- First Published: 17 April 2014
Locus-Specific Databases in Cancer: What Future in a Post-Genomic Era? The TP53 LSDB paradigm
- Pages: 643-653
- First Published: 29 January 2014
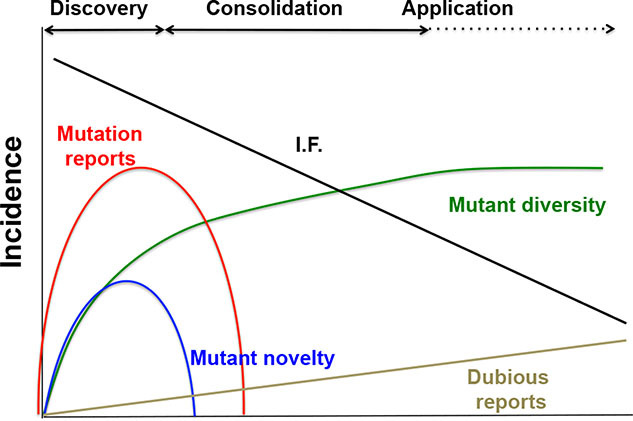
The discovery of a novel gene lead to the discovery phase with an immediate increase in the number of studies leading to the identification of novel mutants (blue line) and mutant diversity rises quickly (green line). This is associated with a high frequency of published reports (red line) in high ranking journals (black line). During the consolidation phase, mutant diversity reaches a plateau the number of mutation reports drops with a decrease in publication quality and an increase in inconsistent reports (brown lines). The application phase will begin only if the gene has clinical value.
Germline TP53 Mutations and the Changing Landscape of Li–Fraumeni Syndrome
- Pages: 654-662
- First Published: 04 April 2014
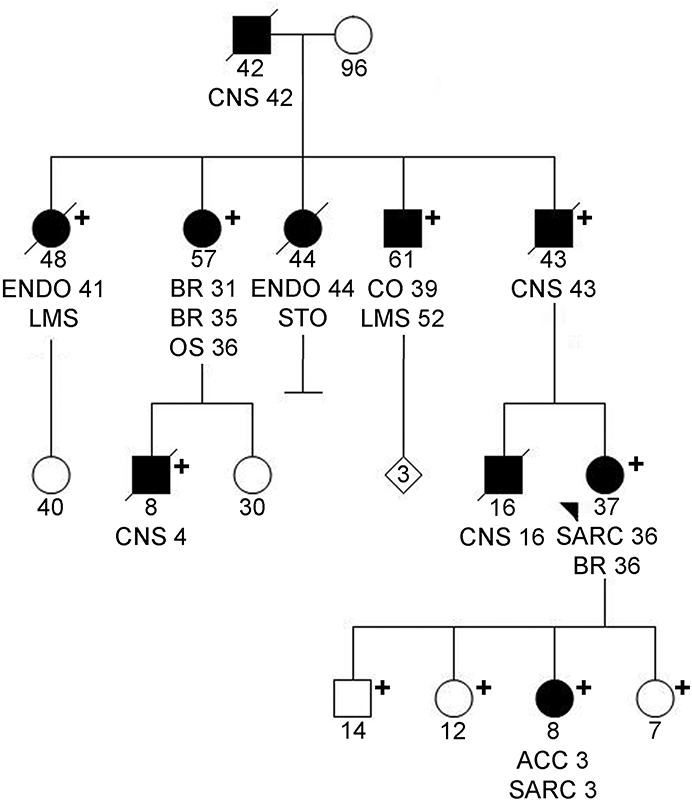
This review highlights Li–Fraumeni syndrome, the familial cancer predisposition syndrome that affects both children and adults, and its association with germline TP53 mutations. Classic and evolving definitions of the syndrome are discussed, highlighting the potential implications that multi-gene analysis may have in expanding our understanding of how phenotype and genotype correlate. Newer options for clinical management of individuals at risk are also discussed.
TP53 Mutation Analysis in Clinical Practice: Lessons From Chronic Lymphocytic Leukemia
- Pages: 663-671
- First Published: 10 January 2014
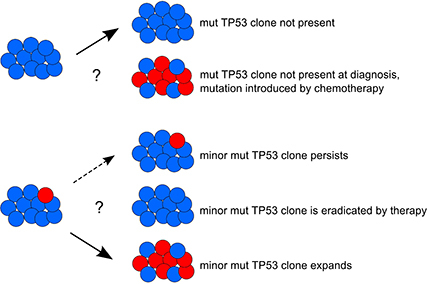
In Chronic lymphocytic leukemia, TP53 defects associate with impaired survival and therapy resistance. Here we describe the biological and clinical consequences of TP53 dysfunction as well as the methodical aspects of TP53 analysis including next generation sequencing. We also discuss the mechanisms of TP53 defects' clonal evolution mainly with respect to their selection by therapy.
TP53 Mutations in Human Cancer: Database Reassessment and Prospects for the Next Decade
- Pages: 672-688
- First Published: 24 March 2014
TP53 Mutants in the Tower of Babel of Cancer Progression
- Pages: 689-701
- First Published: 21 January 2014
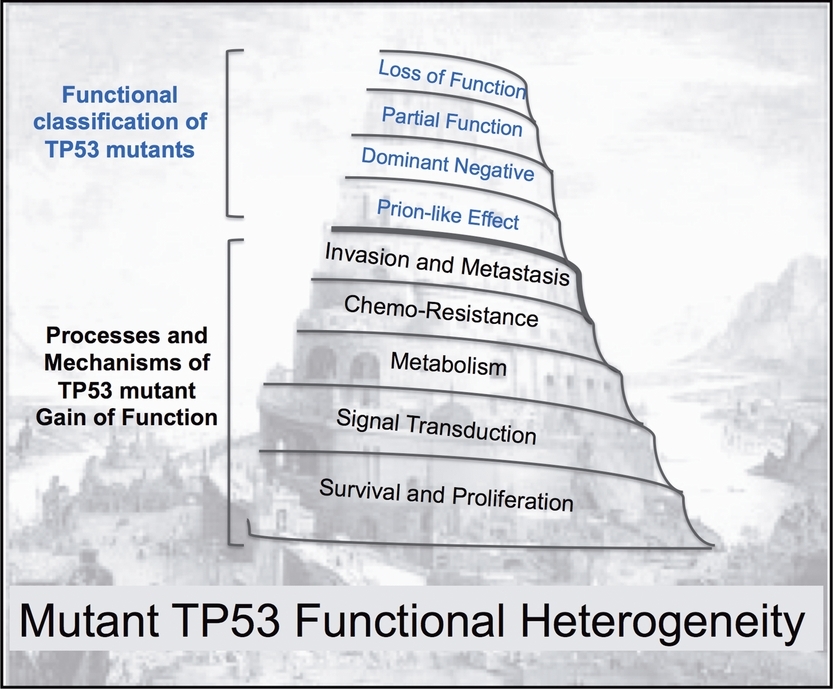
We review studies investigating TP53 mutations for their effect on the capacity of this protein to act as a transcription factor. We also review recent findings on how TP53 mutants can exhibit gain-of-function properties. Somatic as well as germline TP53 mutations are considered, and the possibilities to stratify patients based on the features of P53 mutant proteins are discussed. In the analogy with the Tower of Babel, the different languages stand for the staggering functional heterogeneity of P53 mutants associated with cancer.
How the TP53 Family Proteins TP63 and TP73 Contribute to Tumorigenesis: Regulators and Effectors
- Pages: 702-714
- First Published: 01 February 2014
Insights into Wild-Type and Mutant p53 Functions Provided by Genetically Engineered Mice
- Pages: 715-727
- First Published: 10 January 2014
The TP53 tumor suppressor gene is the most frequently mutated gene in human cancers. Genetically engineered alterations of the Trp53 gene in the mouse germ line have been particularly useful in furthering our understanding of the functions of both the wildtype and mutant forms of p53 at the organismal level.
The Mdm Network and Its Regulation of p53 Activities: A Rheostat of Cancer Risk
- Pages: 728-737
- First Published: 01 February 2014
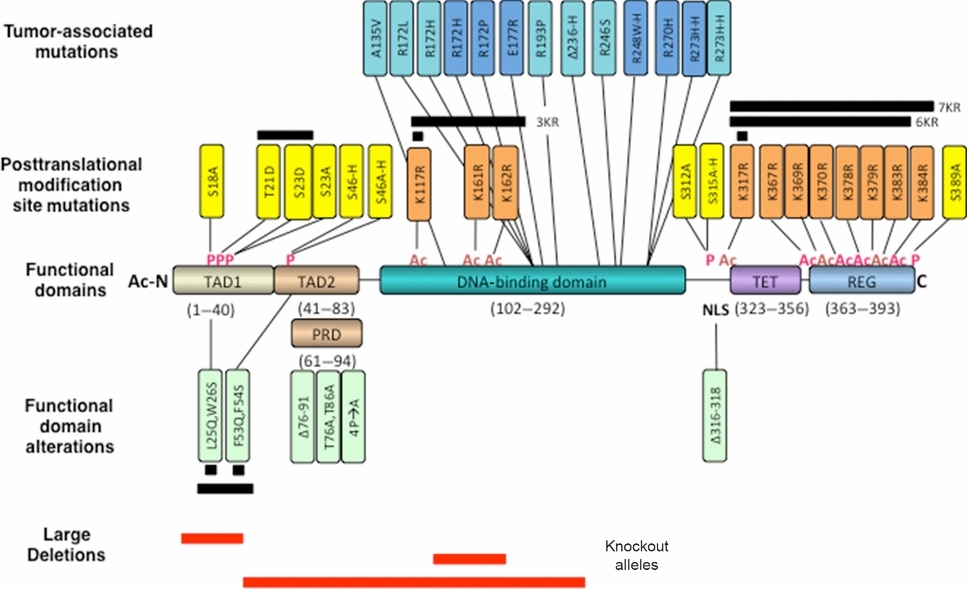
Mutant TP53 Posttranslational Modifications: Challenges and Opportunities
- Pages: 738-755
- First Published: 06 January 2014
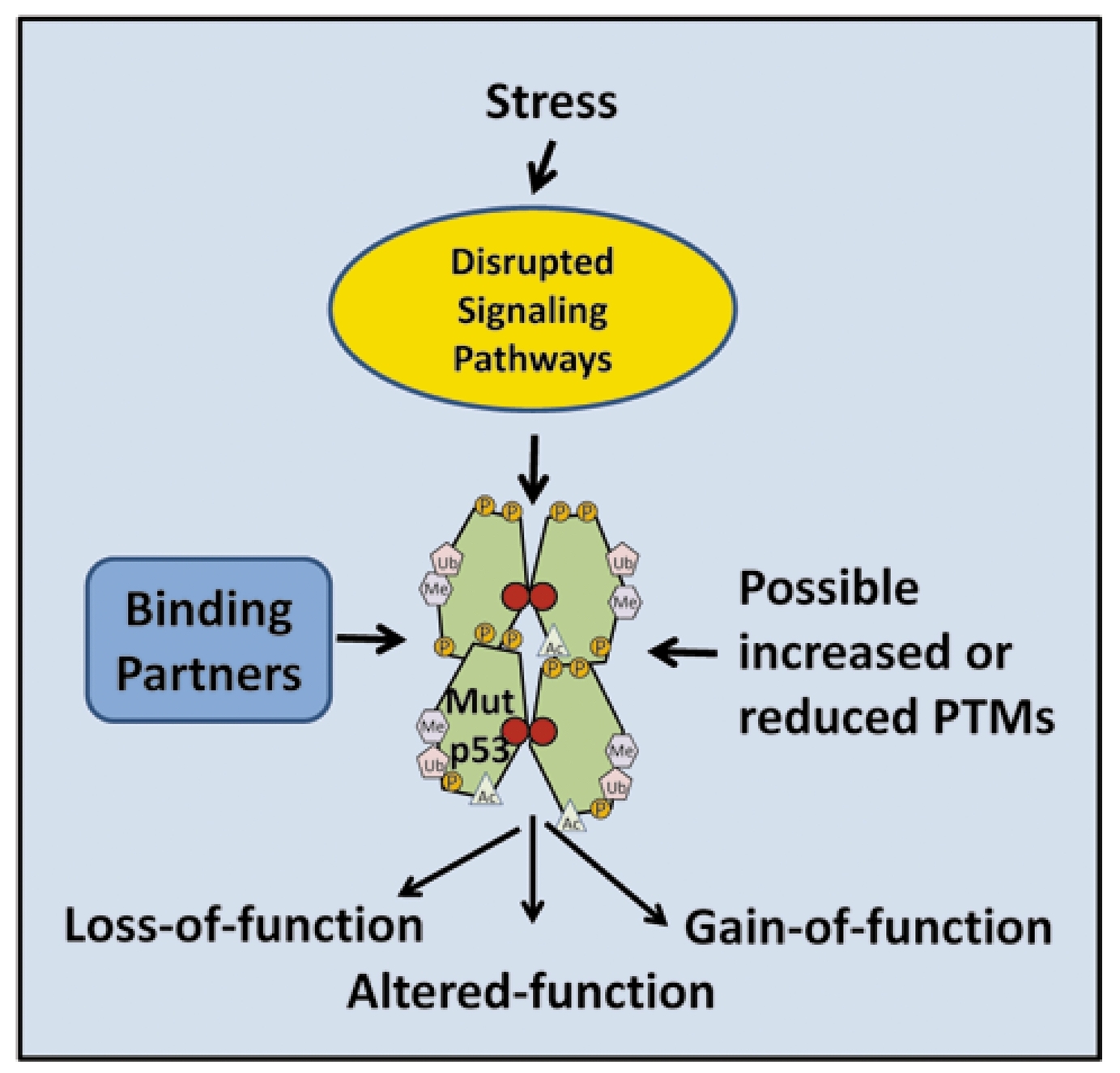
Over half of human cancers contain p53 with a missense mutation. Posttranslational modification appears to occur at a common set of over 60 residues in WT and MUT TP53. Since several stress signaling pathways that modify p53 can be dysregulated in human cancers and since MUT p53 may accumulate to high levels, posttranslational modifications may alter MUT TP53 activities including gain-of-function, thereby influencing tumor formation and metastasis.
Analysis of TP53 Mutation Status in Human Cancer Cell Lines: A Reassessment
- Pages: 756-765
- First Published: 02 April 2014
We have used the 2014 release of the UMD TP53 database to show that TP53 status is still controversial for numerous cell lines, including some widely used lines from the NCI-60 panel. Our analysis clearly confirms that, despite numerous warnings, the misidentification of cell lines is still present as a silent and neglected issue, and that extreme care must be taken when determining the status of p53, because errors may lead to disastrous experimental interpretations. A novel compendium gathering the TP53 status of 2,500 cell lines has been made available (http://p53.fr).
Recommendations for Analyzing and Reporting TP53 Gene Variants in the High-Throughput Sequencing Era
- Pages: 766-778
- First Published: 12 April 2014
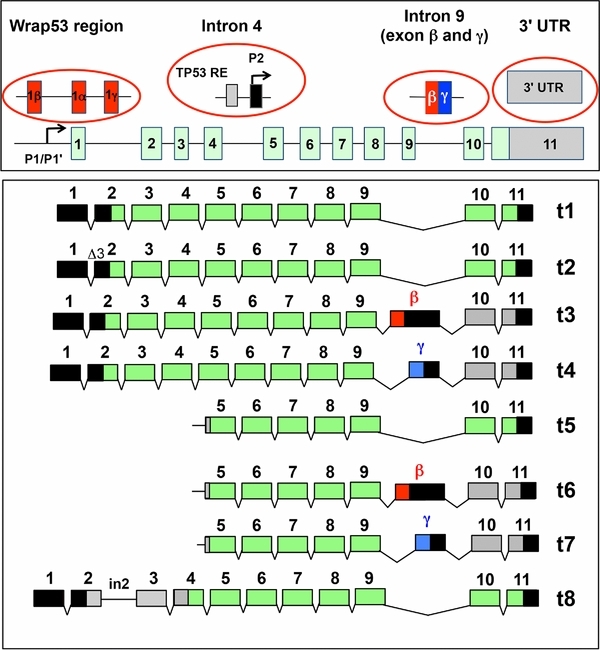
TP53 gene screening in the post-genomic era. Four novel regions of the TP53 gene must be included in screens for alterations to define their importance in tumorigenesis; i: the WRAP53 region that includes overlapping exons with the TP53 gene; ii: intron 4 with a TP53 response element (TP53 RE) and the P2 promoter expressing transcripts t5, t6 and t7; iii: intron 9 with the two novel exons β and γ; iv: the 3' UTR region containing sequences targeted by microRNA mir-125b.
The TP53 gene is transcribed into 8 different mRNAs. Transcripts t1 to t4 originate from promoter P1 and P1' localized upstream of the gene. Transcripts t5 to t8 originate from promoter P2 localized in intron 4.