Journal list menu
Export Citations
Download PDFs
ERF-related craniosynostosis: The phenotypic and developmental profile of a new craniosynostosis syndrome
- First Published: 13 February 2019
Sex differences in the genetic architecture of obsessive–compulsive disorder
- First Published: 20 November 2018
Genomics of body fat percentage may contribute to sex bias in anorexia nervosa
- First Published: 28 December 2018
International multidisciplinary collaboration toward an annotated definition of arthrogryposis multiplex congenita
- First Published: 07 July 2019
Epigenetics and genomics in Turner syndrome
- First Published: 27 February 2019
Association of polymorphisms in genes coding for antioxidant enzymes and human male infertility
- First Published: 07 September 2018
DYNC1H1 gene methylation correlates with severity of spinal muscular atrophy
- First Published: 24 September 2018
The National Spina Bifida Patient Registry: A Decade's journey
- First Published: 06 November 2018
Smoking and pregnancy: Epigenetics and developmental origins of the metabolic syndrome
- First Published: 16 July 2019
Generalizing polygenic risk scores from Europeans to Hispanics/Latinos
- First Published: 15 October 2018
The eMERGE genotype set of 83,717 subjects imputed to ~40 million variants genome wide and association with the herpes zoster medical record phenotype
- First Published: 08 October 2018
Variable population prevalence estimates of germline TP53 variants: A gnomAD-based analysis
- First Published: 23 October 2018
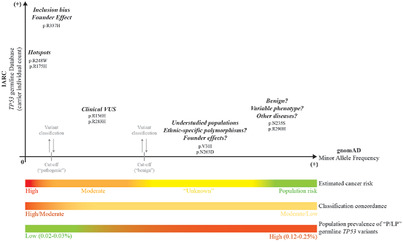
gnomAD-based analysis suggests that the population prevalence of pathogenic and likely pathogenic germline TP53 variants range from 1 carrier in 400 to 5,500 individuals due to differences in variant classification. These estimates may not necessarily reflect the prevalence of Li-Fraumeni syndrome (LFS), which is based upon family history of cancer. We encourage comprehensive approaches to better understand the interplay of germline TP53 variant classification, prevalence estimates, cancer penetrance, and LFS-associated phenotypes.
SLC35A2-CDG: Functional characterization, expanded molecular, clinical, and biochemical phenotypes of 30 unreported Individuals
- First Published: 28 February 2019
Evaluation of the detection of GBA missense mutations and other variants using the Oxford Nanopore MinION
- First Published: 13 January 2019
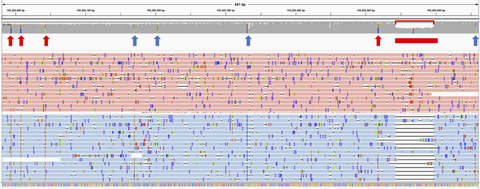
The GBA gene is important in Parkinson's and Gaucher disease, but difficult to sequence due to a highly homologous adjacent pseudogene. Here we present a novel method using long reads on the Oxford Nanopore MinION, which can detect missense mutations and an exonic deletion, with the added advantage of phasing and intronic analysis.
Identification of genes associated with cancer progression and prognosis in lung adenocarcinoma: Analyses based on microarray from Oncomine and The Cancer Genome Atlas databases
- First Published: 16 December 2018
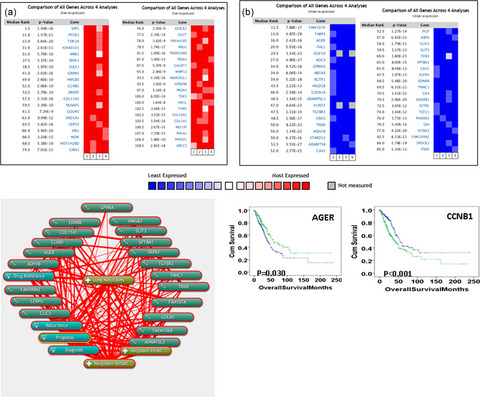
Oncomine and The Cancer Genome Atlas (TCGA) database were useful tools in identification of dysregulated genes in cancers. Twenty consistently and stably dysregulated genes in lung adenocarcinoma (LUAD) were identified based on four microarrays from Oncomine database. AGER and CCNB1 expression were further explored to be associated with survival in LUAD based on TCGA database.



