STED nanoscopy of KK114-stained pathogenic bacteria
Massimiliano Lucidi
Department of Engineering, University Roma Tre, Rome, Italy
Search for more papers by this authorRadu Hristu
Center for Microscopy - Microanalysis and Information Processing, University Politehnica of Bucharest, Bucharest, Romania
Search for more papers by this authorLorenzo Nichele
Department of Engineering, University Roma Tre, Rome, Italy
Search for more papers by this authorGeorge A. Stanciu
Center for Microscopy - Microanalysis and Information Processing, University Politehnica of Bucharest, Bucharest, Romania
Search for more papers by this authorDenis E. Tranca
Center for Microscopy - Microanalysis and Information Processing, University Politehnica of Bucharest, Bucharest, Romania
Search for more papers by this authorAlina Maria Holban
Microbiology and Immunology Department, Faculty of Biology, University of Bucharest, Bucharest, Romania
Search for more papers by this authorPaolo Visca
Department of Sciences, University Roma Tre, Rome, Italy
Search for more papers by this authorCorresponding Author
Stefan G. Stanciu
Center for Microscopy - Microanalysis and Information Processing, University Politehnica of Bucharest, Bucharest, Romania
Correspondence
Stefan G. Stanciu, Center for Microscopy - Microanalysis and Information Processing, University Politehnica of Bucharest, 313 Splaiul Independentei, 060042 Bucharest, Romania.
Email: [email protected]
Gabriella Cincotti, Department of Engineering, University Roma Tre, via Vito Volterra 62, 00146 Rome, Italy.
Email: [email protected]
Search for more papers by this authorCorresponding Author
Gabriella Cincotti
Department of Engineering, University Roma Tre, Rome, Italy
Correspondence
Stefan G. Stanciu, Center for Microscopy - Microanalysis and Information Processing, University Politehnica of Bucharest, 313 Splaiul Independentei, 060042 Bucharest, Romania.
Email: [email protected]
Gabriella Cincotti, Department of Engineering, University Roma Tre, via Vito Volterra 62, 00146 Rome, Italy.
Email: [email protected]
Search for more papers by this authorMassimiliano Lucidi
Department of Engineering, University Roma Tre, Rome, Italy
Search for more papers by this authorRadu Hristu
Center for Microscopy - Microanalysis and Information Processing, University Politehnica of Bucharest, Bucharest, Romania
Search for more papers by this authorLorenzo Nichele
Department of Engineering, University Roma Tre, Rome, Italy
Search for more papers by this authorGeorge A. Stanciu
Center for Microscopy - Microanalysis and Information Processing, University Politehnica of Bucharest, Bucharest, Romania
Search for more papers by this authorDenis E. Tranca
Center for Microscopy - Microanalysis and Information Processing, University Politehnica of Bucharest, Bucharest, Romania
Search for more papers by this authorAlina Maria Holban
Microbiology and Immunology Department, Faculty of Biology, University of Bucharest, Bucharest, Romania
Search for more papers by this authorPaolo Visca
Department of Sciences, University Roma Tre, Rome, Italy
Search for more papers by this authorCorresponding Author
Stefan G. Stanciu
Center for Microscopy - Microanalysis and Information Processing, University Politehnica of Bucharest, Bucharest, Romania
Correspondence
Stefan G. Stanciu, Center for Microscopy - Microanalysis and Information Processing, University Politehnica of Bucharest, 313 Splaiul Independentei, 060042 Bucharest, Romania.
Email: [email protected]
Gabriella Cincotti, Department of Engineering, University Roma Tre, via Vito Volterra 62, 00146 Rome, Italy.
Email: [email protected]
Search for more papers by this authorCorresponding Author
Gabriella Cincotti
Department of Engineering, University Roma Tre, Rome, Italy
Correspondence
Stefan G. Stanciu, Center for Microscopy - Microanalysis and Information Processing, University Politehnica of Bucharest, 313 Splaiul Independentei, 060042 Bucharest, Romania.
Email: [email protected]
Gabriella Cincotti, Department of Engineering, University Roma Tre, via Vito Volterra 62, 00146 Rome, Italy.
Email: [email protected]
Search for more papers by this authorFunding information: European Cooperation in Science and Technology, Grant/Award Number: CA17121 COMULIS; European Regional Development Fund, Grant/Award Number: P_36_611; MySMIS code 107066; Unitatea Executiva pentru Finantarea Invatamantului Superior, a Cercetarii, Dezvoltarii si Inovarii, Grant/Award Number: PN-III-P1-1.1-TE-2016-2147 CORIMAG
Abstract
Super-resolution microscopy techniques can provide answers to still pending questions on prokaryotic organisms but are yet to be used at their full potential for this purpose. To address this, we evaluate the ability of the rhodamine-like KK114 dye to label various types of bacteria, to enable imaging of fine structural details with stimulated emission depletion microscopy (STED). We assessed fluorescent labeling with KK114 for eleven Gram-positive and Gram-negative bacterial species and observed that this contrast agent binds to their cell membranes. Significant differences in the labeling outputs were noticed across the tested bacterial species, but importantly, KK114-staining allowed the observation of subtle nanometric cell details in some cases. For example, a helix pattern resembling a cytoskeleton arrangement was detected in Bacillus subtilis. Furthermore, we found that KK114 easily penetrates the membrane of bacterial microorganism that lost their viability, which can be useful to discriminate between living and dead cells.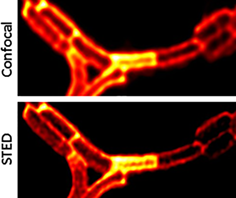
CONFLICT OF INTEREST
The authors declare no financial or commercial conflict of interest.
REFERENCES
- 1 Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics,” WHO. [Online]. Available: http://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/.
- 2L. B. Rice, J. Infect. Dis. 2008, 197, 1079.
- 3S. Santajit, N. Indrawattana, Biomed. Res. Int. 2016, 2016, 2475067.
- 4M. J. Franklin, C. Chang, T. Akiyama, B. Bothner, Microbiol. Spectr. 2015, 3, 1.
- 5F. Dempwolff, F. K. Schmidt, A. B. Hervás, A. Stroh, T. C. Rösch, C. N. Riese, S. Dersch, T. Heimerl, D. Lucena, N. Hülsbusch, C. A. O. Stuermer, N. Takeshita, R. Fischer, B. Eckhardt, P. L. Grauman, PLoS Genet. 2016, 12, e1006116.
- 6K. D. Young, Microbiol. Mol. Biol. Rev. 2006, 70, 660.
- 7M. C. F. van Teeseling, M. A. de Pedro, F. Cava, Front. Microbiol. 2017, 8, 1264.
- 8D. Du, X. Wang-Kan, A. Neuberger, H. W. van Veen, K. M. Pos, L. J. V. Piddock, B. F. Luisi, Nat. Rev. Microbiol. 2018, 16, 523.
- 9L. Pasquina-Lemonche, J. Burns, R. D. Turner, et al. The architecture of the Gram-positive bacterial cell wall, Nature 2020, https://doi.org/10.1038/s41586-020-2236-6.
- 10L. Schermelleh, R. Heintzmann, H. Leonhardt, J. Cell, Biol. 2010, 190, 165.
- 11G. E. Fantner, R. J. Barbero, D. S. Gray, A. M. Belcher, Nat. Nanotechnol. 2010, 5, 280.
- 12A. Razatos, Y. L. Ong, M. M. Sharma, G. Georgiou, Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 11059.
- 13P. Hildebrandt, K. Surmann, M. G. Salazar, N. Normann, U. Völker, F. Schmidt, Cytometry A 2016, 89, 932.
- 14D. A. Drevets, A. M. Elliott, J. Immunol. Methods 1995, 187, 69.
- 15J. P. Schneider, M. Basler, Philos. Trans. R. Soc. 2016, 371, 0150499.
- 16S. J. Sahl, S. W. Hell, S. Jakobs, Nat. Rev. Mol. Cell Biol. 2017, 18, 685.
- 17M. Heilemann, J. Biotechnol. 2010, 149, 243.
- 18C. Polzer, S. Ness, M. Mohseni, T. Kellerer, M. Hilleringmann, J. Rädler, T. Hellerer, Biomed. Opt. Express 2019, 10, 4516.
- 19A. Gahlmann, W. E. Moerner, Nat. Rev. Microbiol. 2014, 12, 9.
- 20R. Wheeler, S. Mesnage, I. G. Boneca, J. K. Hobbs, S. J. Foster, Mol. Microbiol. 2011, 82, 1096.
- 21S. Saurabh, A. M. Perez, C. J. Comerci, L. Shapiro, W. E. Moerner, J. Am. Chem. Soc. 2016, 138, 10398.
- 22H. H. Tuson, J. S. Biteen, Anal. Chem. 2015, 87, 42.
- 23S. A. Lee, A. Ponjavic, C. Siv, S. F. Lee, J. S. Biteen, ACS Nano 2016, 10, 8143.
- 24G. Vicidomini, P. Bianchini, A. Diaspro, Nat. Methods 2018, 15, 173.
- 25C. A. Wurm, K. Kolmakov, F. Göttfert, H. Ta, M. Bossi, H. Schill, S. Berning, S. Jakobs, G. Donnert, V. N. Belov, S. W. Hell, Opt. Nano. 2012, 1, 7.
10.1186/2192-2853-1-7 Google Scholar
- 26C. Spahn, J. B. Grimm, L. D. Lavis, M. Lampe, M. Heilemann, Nano Lett. 2019, 19, 500.
- 27T. Müller, C. Schumann, A. Kraegeloh, ChemPhysChem 2012, 13, 1986.
- 28C. Coltharp, J. Xiao, Cell. Microbiol. 2012, 14, 1808.
- 29K. Cramer, A.-L. Bolender, I. Stockmar, R. Jungmann, R. Kasper, J. Y. Shin, Int. J. Mol. Sci. 2019, 20.
- 30P. C. Jennings, G. C. Cox, L. G. Monahan, E. J. Harry, Micron 2011, 42, 336.
- 31K. Kolmakov, V. N. Belov, J. Bierwagen, C. Ringemann, V. Müller, C. Eggeling, S. W. Hell, Chem. Eur. J. 2010, 16, 158.
- 32J. H. M. van der Velde, J. Oelerich, J. Huang, J. H. Smit, A. A. Jazi, S. Galiani, K. Kolmakov, G. Gouridis, C. Eggeling, A. Herrmann, G. Roelfes, T. Cordes, Nat. Commun. 2016, 7, 10144.
- 33C. Osseforth, J. R. Moffitt, L. Schermelleh, J. Michaelis, Opt. Express 2014, 22, 7028.
- 34P. Nonejuie, M. Burkart, K. Pogliano, J. Pogliano, Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 16169.
- 35D. T. Quach, G. Sakoulas, V. Nizet, J. Pogliano, K. Pogliano, EBioMedicine 2016, 4, 95.
- 36H. H. Htoo, L. Brumage, V. Chaikeeratisak, H. Tsunemoto, J. Sugie, C. Tribuddharat, J. Pogliano, P. Nonejuie, Antimicrob. Agents Chemother. 2019, 63, e02310-18, 1.
- 37P. Sass, H. Brötz-Oesterhelt, Curr. Opin. Microbiol. 2013, 16, 522.
- 38C. Schaffner-Barbero, M. Martín-Fontecha, P. Chacón, J. M. Andreu, ACS Chem. Biol. 2012, 7, 269.
- 39S. Pajk, H. Majaron, M. Novak, B. Kokot, J. Štrancar, Eur. Biophys. J. 2019, 48, 485.
- 40Z. Huang, E. London, Chem. Phys. Lipids 2016, 199, 11.
- 41A. P. A. Hendrickx, W. van Schaik, R. J. L. Willems, Future Microbiol. 2013, 8, 993.
- 42J. M. Monteiro, P. B. Fernandes, F. Vaz, A. R. Pereira, A. C. Tavares, M. T. Ferreira, P. M. Pereira, H. Veiga, E. Kuru, M. S. VanNieuwenhze, Y. V. Brun, S. R. Filipe, M. G. Pinho, Nat. Commun. 2015, 6, 8005.
- 43A. Honigmann, V. Mueller, S. W. Hell, C. Eggeling, Faraday Discuss. 2013, 161, 77.
- 44S. K. Saka, A. Honigmann, C. Eggeling, S. W. Hell, T. Lang, S. O. Rizzoli, Nat. Commun. 2014, 5, 4509.
- 45L. G. Monahan, A. Robinson, E. J. Harry, Mol. Microbiol. 2009, 74, 1004.
- 46L. J. Jones, R. Carballido-López, J. Errington, Cell 2001, 104, 913.
- 47S. H. Ho, D. A. Tirrell, ACS Cent. Sci. 2019, 5, 1911.
- 48A. J. Hyde, J. Parisot, A. McNichol, B. B. Bonev, Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 19896.
- 49D. J. Haydon, N. R. Stokes, R. Ure, G. J. M. B. Galbraith, D. R. Brown, P. J. Baker, V. V. Barynin, D. W. Rice, S. E. Sedelnikova, J. R. Heal, J. M. Sheridan, S. T. Aiwale, P. K. Chauhan, A. Srivastava, A. Taneja, I. Collins, J. Errington, L. G. Czaplewski, Science 2008, 321, 1673.
- 50T. J. Silhavy, D. Kahne, S. Walker, Cold Spring Harb. Perspect. Biol. 2010, 2, a000414.
- 51N. Baranova, M. Loose, Methods Cell Biol. 2017, 137, 355.
- 52P. H. Lebre, P. De Maayer, D. A. Cowan, Nat. Rev. Microbiol. 2017, 15, 285.
- 53M. Berney, F. Hammes, F. Bosshard, H.-U. Weilenmann, T. Egli, Appl. Environ. Microbiol. 2007, 73, 3283.
- 54L. Yang, Y. Zhou, S. Zhu, T. Huang, L. Wu, X. Yan, Anal. Chem. 2012, 84, 1526.
- 55K. Lewis, Microbiol. Mol. Biol. Rev. 2000, 64, 503.
- 56R. L. Soon, R. L. Nation, P. G. Hartley, I. Larson, J. Li, Antimicrob. Agents Chemother. 2009, 53, 4979.




