Journal list menu
Export Citations
Download PDFs
editorial
Submission of structural biology data for review purposes
- Pages: 1477-1478
- First Published: 06 December 2021
research papers
Correlative cryo-imaging of the cellular universe with soft X-rays and laser light used to track F-actin structures in mammalian cells
- Pages: 1479-1485
- First Published: 30 November 2021
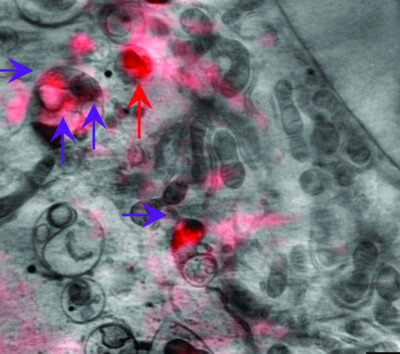
A 3D cellular imaging platform developed at beamline B24 at Diamond Light Source has been used to identify cellular features such as filamentous actin in mammalian cells. This approach enabled virtual same-sample imaging of structures captured on separate microscopes through rigid transformation of 3D data in silico, bypassing the need for additional sample processing and ensuring artefact-free data correlation.
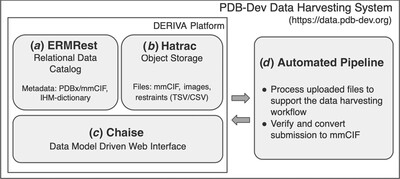
New system for archiving integrative structures
- Pages: 1486-1496
- First Published: 30 November 2021
Ten things I `hate' about refinement
- Pages: 1497-1515
- First Published: 30 November 2021
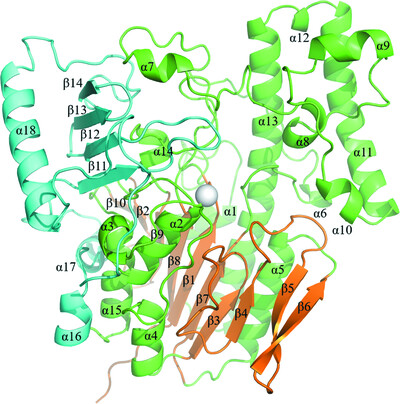
Structural and thermodynamic insights into a novel Mg2+–citrate-binding protein from the ABC transporter superfamily
- Pages: 1516-1534
- First Published: 11 November 2021
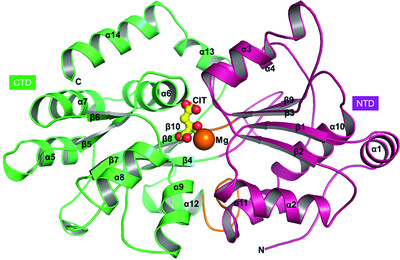
MctA is an Mg2+-complexed citrate-binding protein from a Gram-negative bacterium that belongs to the ABC transporter superfamily. Comparison of the crystal structures of wild-type and mutant MctA proteins suggest a gating mechanism of substrate entry following an `asymmetric domain movement' mechanism of substrate binding.
Legionella effector LegA15/AnkH contains an unrecognized cysteine protease-like domain and displays structural similarity to LegA3/AnkD, but differs in host cell localization
- Pages: 1535-1542
- First Published: 11 November 2021
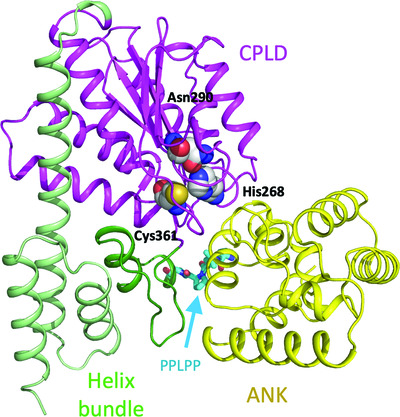
Legionella pneumophila protein effector LegA15/AnkD contains an ankyrin-repeat domain, a cysteine protease-like domain with His268–Asn290–Cys361 putative catalytic triad, and a helix-bundle domain. LegA15/AnkD shows structural similarity to another effector, LegA3/AnkH, but they localize to different cellular compartment within the host.
Helical filament structure of the DREP3 CIDE domain reveals a unified mechanism of CIDE-domain assembly
- Pages: 1543-1553
- First Published: 11 November 2021
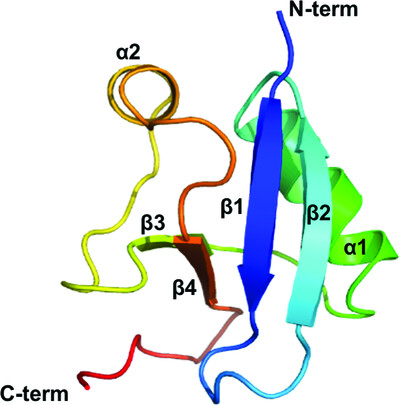
The CIDE domain was initially identified in apoptotic nucleases and now forms a highly conserved family with diverse functions ranging from cell death to lipid metabolism. Based on structural determination of the DREP3 domain, it is suggested that the head-to-tail helical filament structure might be a unified mechanism of CIDE-domain assembly and represents a critical higher-order scaffolding structure that is important for the function of CIDE-domain-containing proteins in DNA fragmentation and lipid-droplet fusion.
Structural and biochemical analyses of the tetrameric carboxypeptidase S9Cfn from Fusobacterium nucleatum
- Pages: 1554-1563
- First Published: 18 November 2021
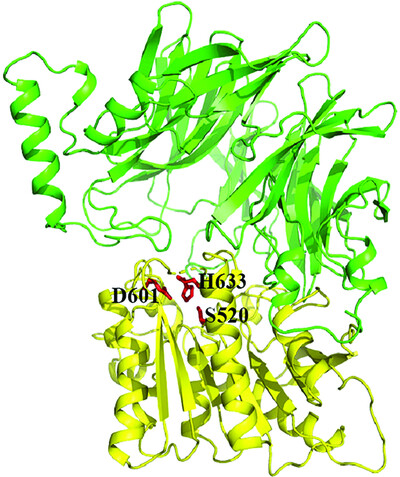
Extensive biochemical assays revealed that a putative peptidase S9Cfn from Fusobacterium nucleatum is a carboxypeptidase rather than an aminopeptidase as previously noted. Combined with the 2.6 Å tetramer structure of S9Cfn, key residues for substrate binding and catalysis, as well as the underlying mechanism of the catalytic cycle was revealed for S9C peptidase family.
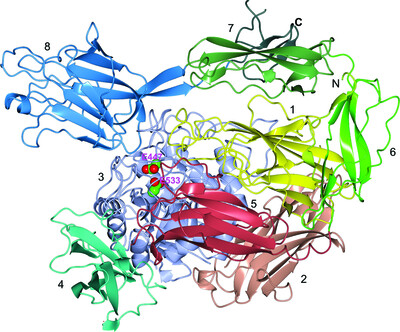
Multitasking in the gut: the X-ray structure of the multidomain BbgIII from Bifidobacterium bifidum offers possible explanations for its alternative functions
- Pages: 1564-1578
- First Published: 18 November 2021
The impact of folding modes and deuteration on the atomic resolution structure of hen egg-white lysozyme
- Pages: 1579-1590
- First Published: 18 November 2021
Predicting the performance of automated crystallographic model-building pipelines
- Pages: 1591-1601
- First Published: 30 November 2021
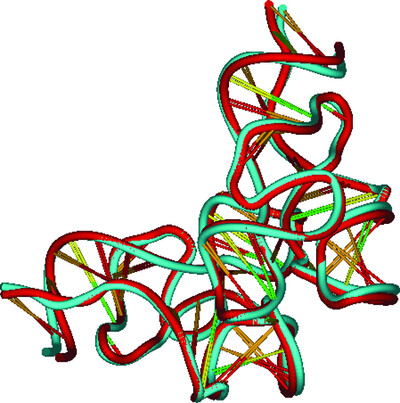
Towards the automatic crystal structure solution of nucleic acids: automated model building using the new CAB program
- Pages: 1602-1613
- First Published: 30 November 2021
Structural analysis of the sulfatase AmAS from Akkermansia muciniphila
- Pages: 1614-1623
- First Published: 30 November 2021