Journal list menu
Export Citations
Download PDFs
ISSUE INFORMATION
ORIGINAL ARTICLES
Assessing the performance of short 18S rDNA markers for environmental DNA metabarcoding of marine protists
- First Published: 30 June 2024
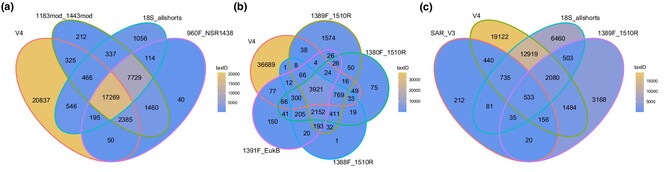
Protists are ubiquitous in aquatic ecosystems, where they perform vital functions. Their diverse nature and limited fossil record make DNA metabarcoding coupled with sedimentary ancient DNA a promising avenue to characterize past ecosystem responses to perturbations. The fragmented state of the DNA requires an informed marker choice, balancing marker length and taxonomic resolution. Using in silico PCR, we offer a comprehensive assessment of short 18S markers and discuss their strengths and weaknesses for sedaDNA studies.
ORIGINAL ARTICLE
A quantitative analysis of vertebrate environmental DNA degradation in soil in response to time, UV light, and temperature
- First Published: 03 July 2024
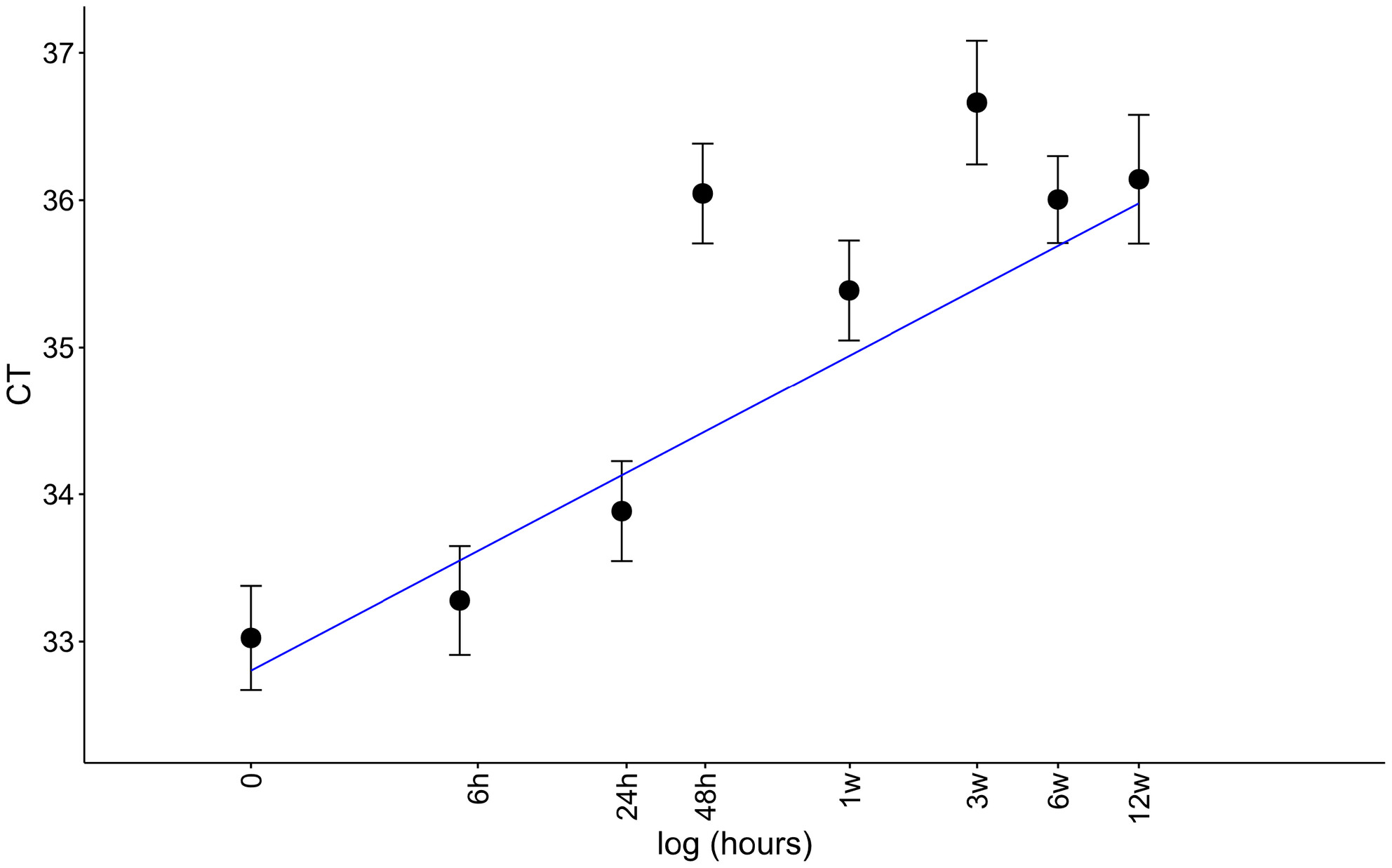
We assessed the persistence of vertebrate eDNA from a mock vertebrate community created with soil from zoo enclosures holding 10 target species from different taxonomic classes (reptiles, birds, and mammals) and of different biomass (little penguin and giraffe). Quantitative polymerase chain reaction indicated degradation of eDNA over time for all temperature and UV treatments, with degradation rates lowest for high UV-B treatments, presumably due to UV-B-reducing bacterial metabolism. Sheltered sites with minimal UV-B radiation, which have previously been considered ideal sites for terrestrial eDNA collection, may not be optimal for eDNA persistence in some cases due to microbial decay, and a better understanding of terrestrial eDNA degradation is needed to improve the accuracy of soil eDNA metabarcoding for surveying of terrestrial vertebrate communities.
ORIGINAL ARTICLES
Environmental DNA for monitoring the impact of offshore wind farms on fish and invertebrate community structures
- First Published: 30 June 2024
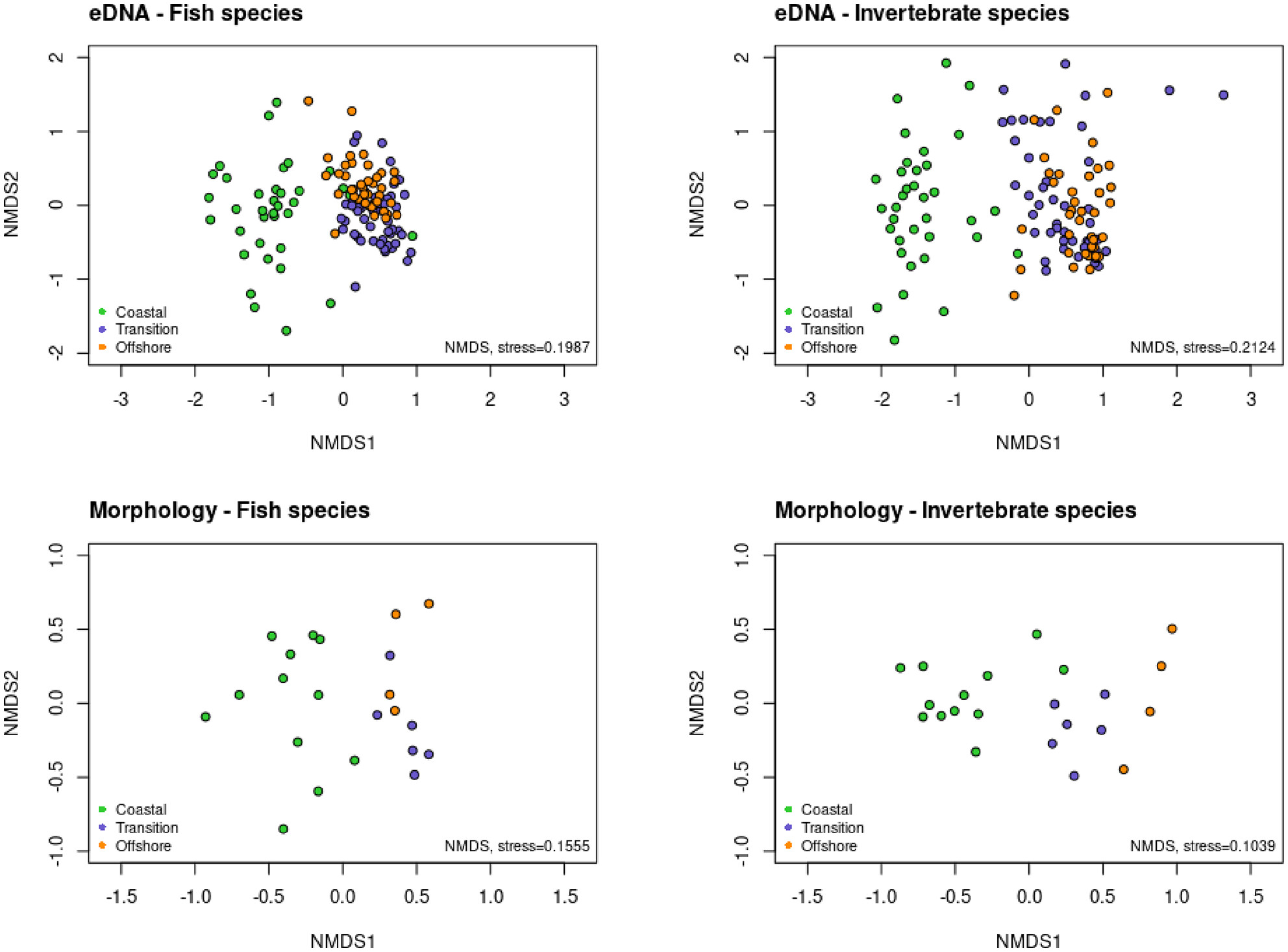
Offshore windfarms are marine areas that are difficult to access, and innovative monitoring techniques are needed to study their impact on marine biodiversity. Water sampling and trawl catches conducted in parallel in 30 sites in the Belgian part of the North Sea showed that 12S eDNA metabarcoding retrieved 85.7% of the fish species caught in the beam trawls, whereas the COI eDNA metabarcoding only identified 31.4% of the epibenthic invertebrate species. Significant differences were found in fish and invertebrate community structures between the coastal, transition and offshore zones as well as on the smaller wind farm scales, which agreed with the morphological beam trawl data. Our findings show that 12S eDNA metabarcoding analyses offers a useful survey tool for the monitoring of fish communities in offshore wind farms.
ORIGINAL ARTICLE
Integration of citizen science and eDNA reveals novel ecological insights for marine fish conservation
- First Published: 09 July 2024
Can environmental DNA be used within pest insect agricultural biosecurity? Detecting khapra beetle within stored rice
- First Published: 09 July 2024
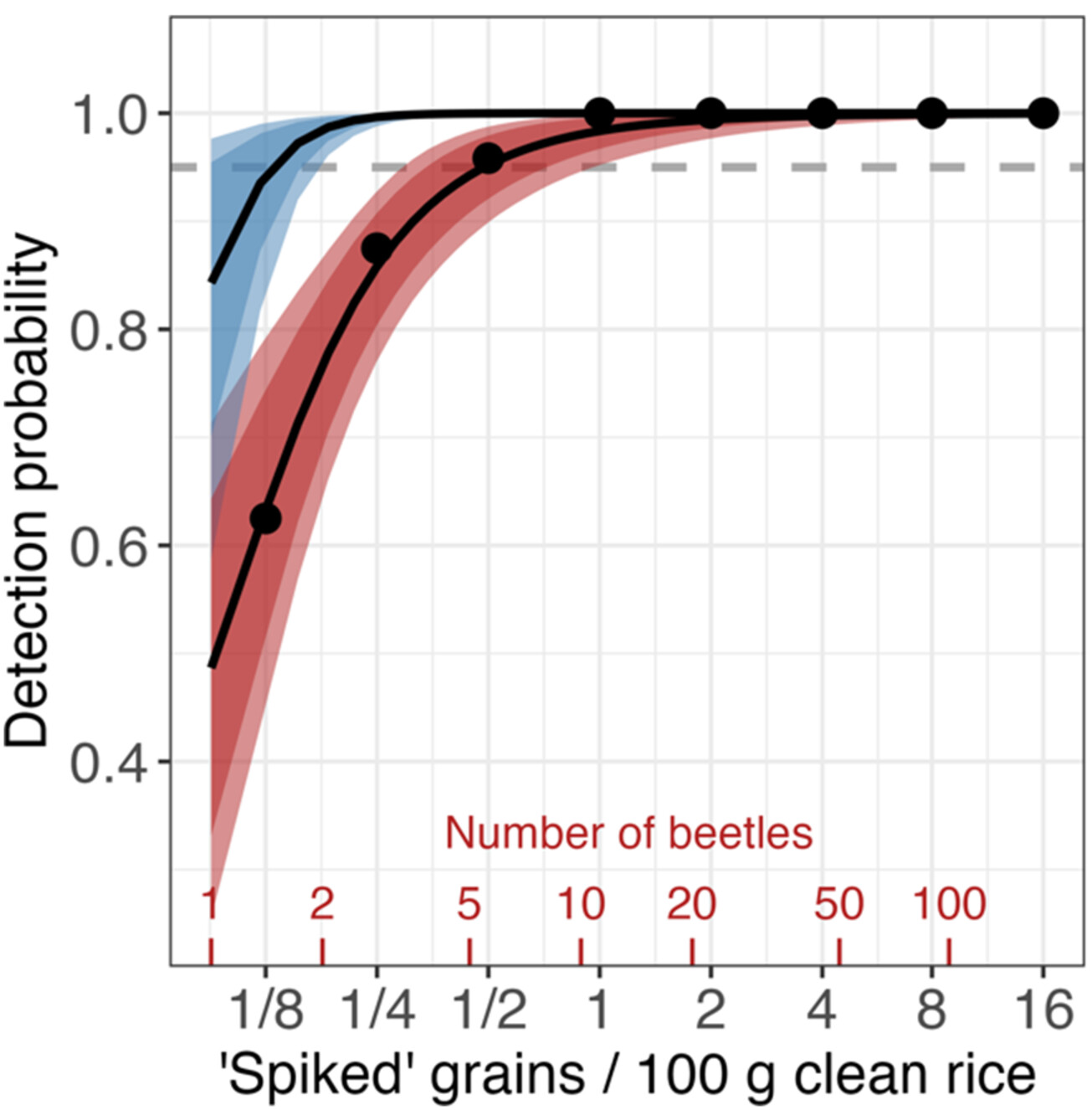
Khapra beetle is a agricultural pest posing high global biosecurity risk. We show eDNA sampling of stored grain can detect ~1 khapra beetle larva in a 50 kg container of rice with 85% to >97% certainty (dashed line is 95% certainty level) depending on whether 1 (red) or 3 (blue) qPCR technical replicates are performed. This result suggests, with further exploration, eDNA surveys can become cost-effective first-step biosecurity protocol for khapra beetle, or a means to map the relative magnitude of khapra beetle transport pathways.
Detection differences between eDNA and mid-water trawls are driven by fish biomass and habitat preferences
- First Published: 14 July 2024
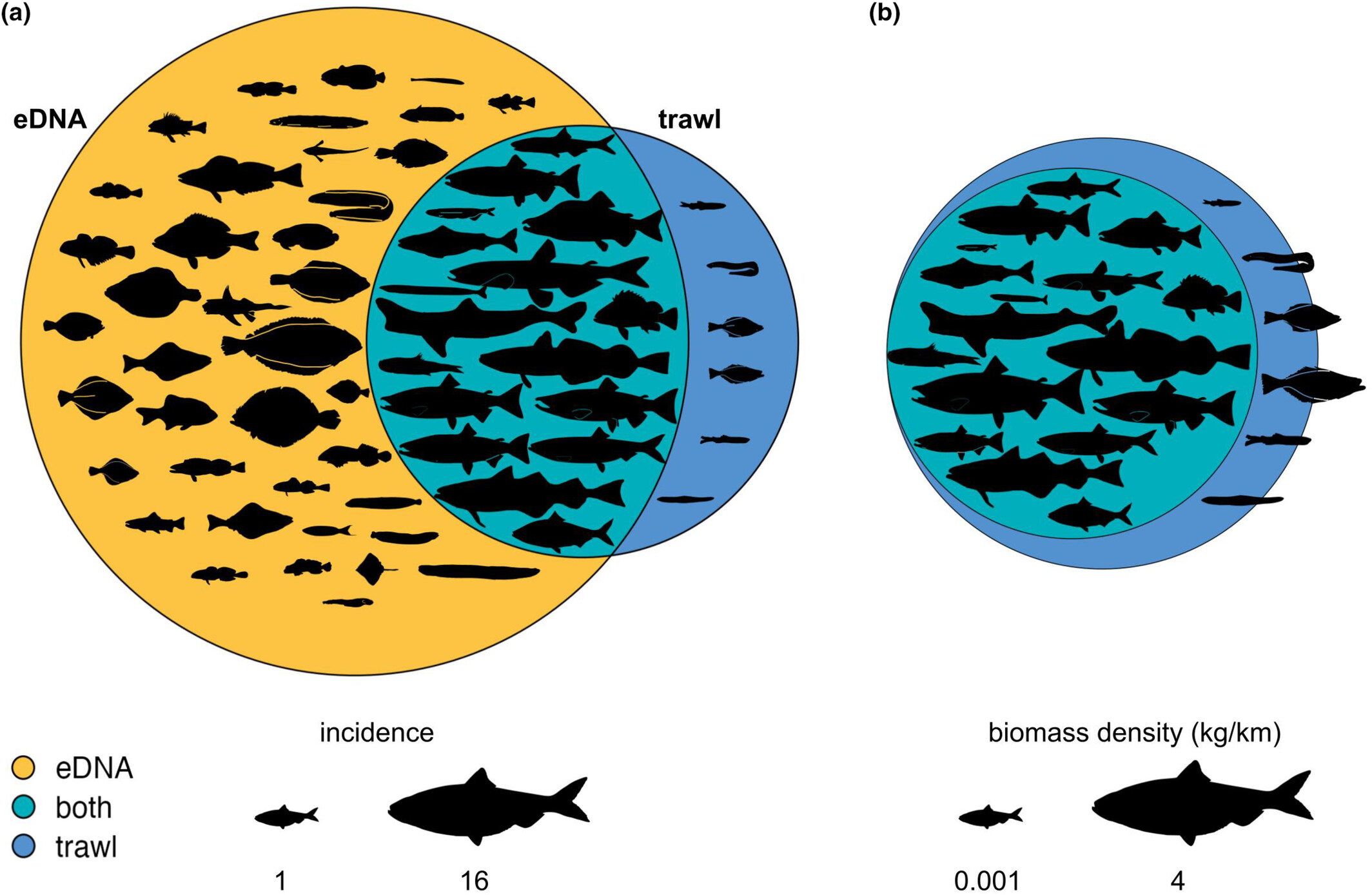
Our study in British Columbia, Canada, reveals that environmental DNA (eDNA) metabarcoding effectively detects fish communities observed by trawl surveys, but with a bias toward species of higher local biomass and demersal habitat preferences. While 74% of trawl-captured species were detected by eDNA, an additional 39 species were exclusively detected through eDNA sampling, indicating its potential for comprehensive species detection beyond traditional methods. Our findings highlight the importance of considering species traits, such as body size and habitat preference, in understanding detection differences between eDNA and trawl surveys, thus advancing our ability to assess marine ecosystems accurately.
Environmental DNA metabarcoding on aquatic insects: Comparing the primer sets of MtInsects-16S based on the mtDNA 16S and general marker based on the mtDNA COI region
- First Published: 18 July 2024
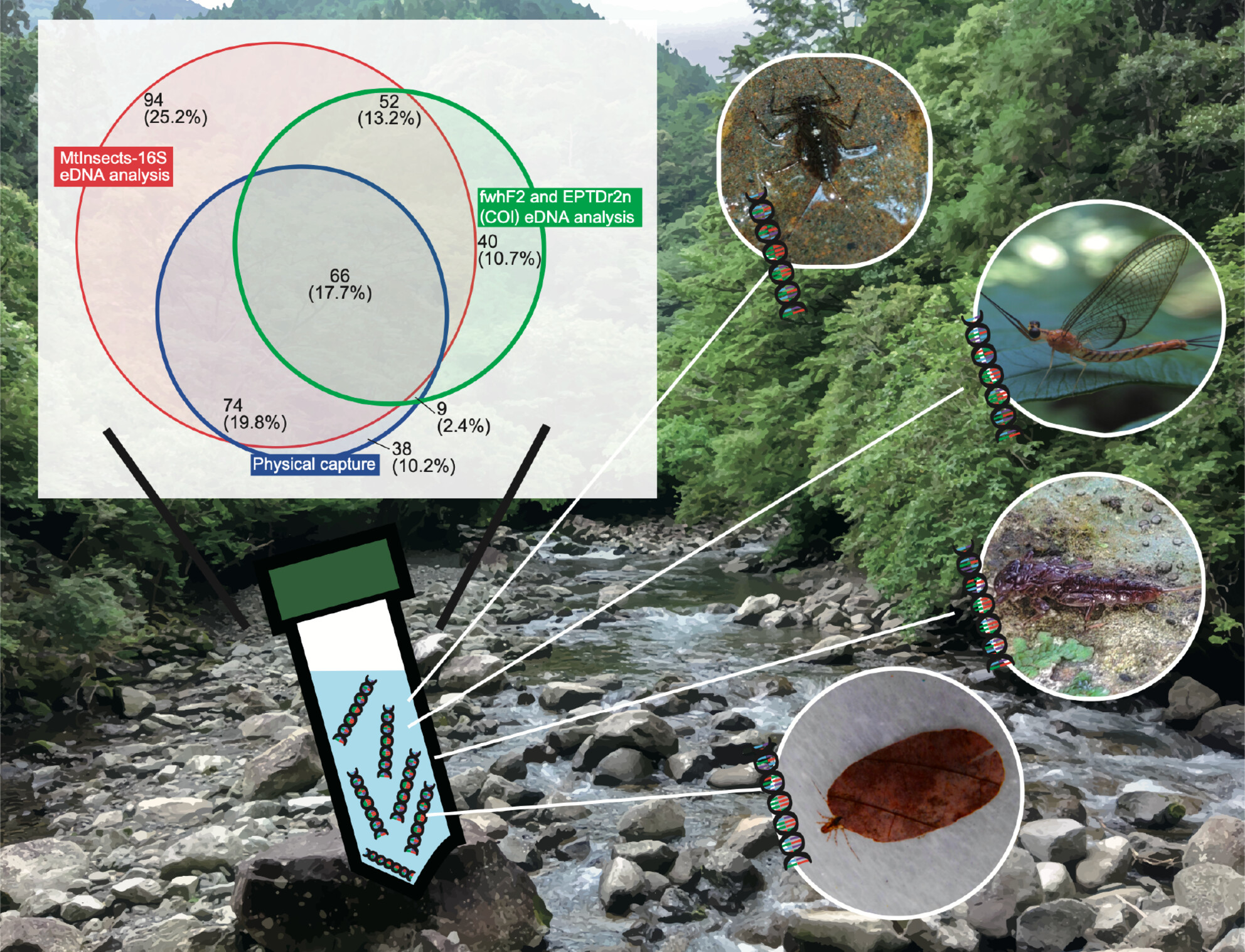
We conducted eDNA analyses using the MtInsects-16S primer set and also compared it with the COI primer set (fwhF2 and EPTDr2n). As a result, eDNA analyses using MtInsects-16S could detected almost all of the captured species (percentage of detected species within collected species by actual capture survey is 87.5%), but using primer set of the mtDNA COI region could not detect nearly half of the captured species (49.3%).
METHOD
Optimization and application of bacterial environmental DNA and RNA isolation for qualitative and quantitative studies
- First Published: 16 July 2024
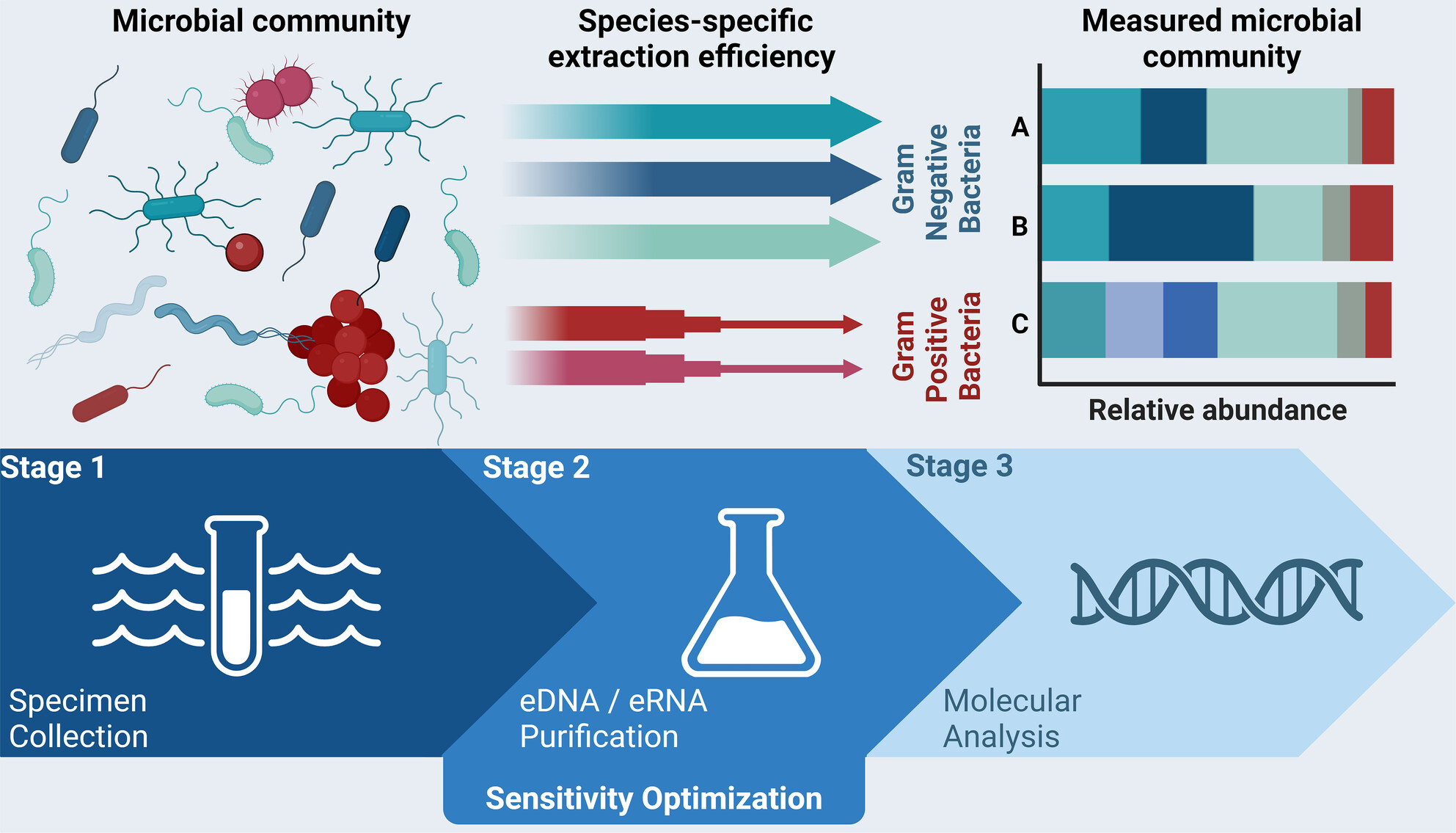
This study investigated the optimal method for isolating bacterial environmental DNA (eDNA) and RNA (eRNA) from aquatic samples by evaluating various filtration and purification systems. When evaluating the analytical sensitivity of the optimized protocol, this study revealed significant species-specific changes in detectable cell number and nucleic acid concentrations that occurred postcollection and identified a bias toward isolation of Gram-negative bacteria. The findings underscore the need for species-specific sensitivity assessments in molecular diagnostics for aquatic bacterial monitoring.
ORIGINAL ARTICLE
Enhancing metabarcoding of freshwater biotic communities: A new online tool for primer selection and exploring data from 14 primer pairs
- First Published: 23 July 2024
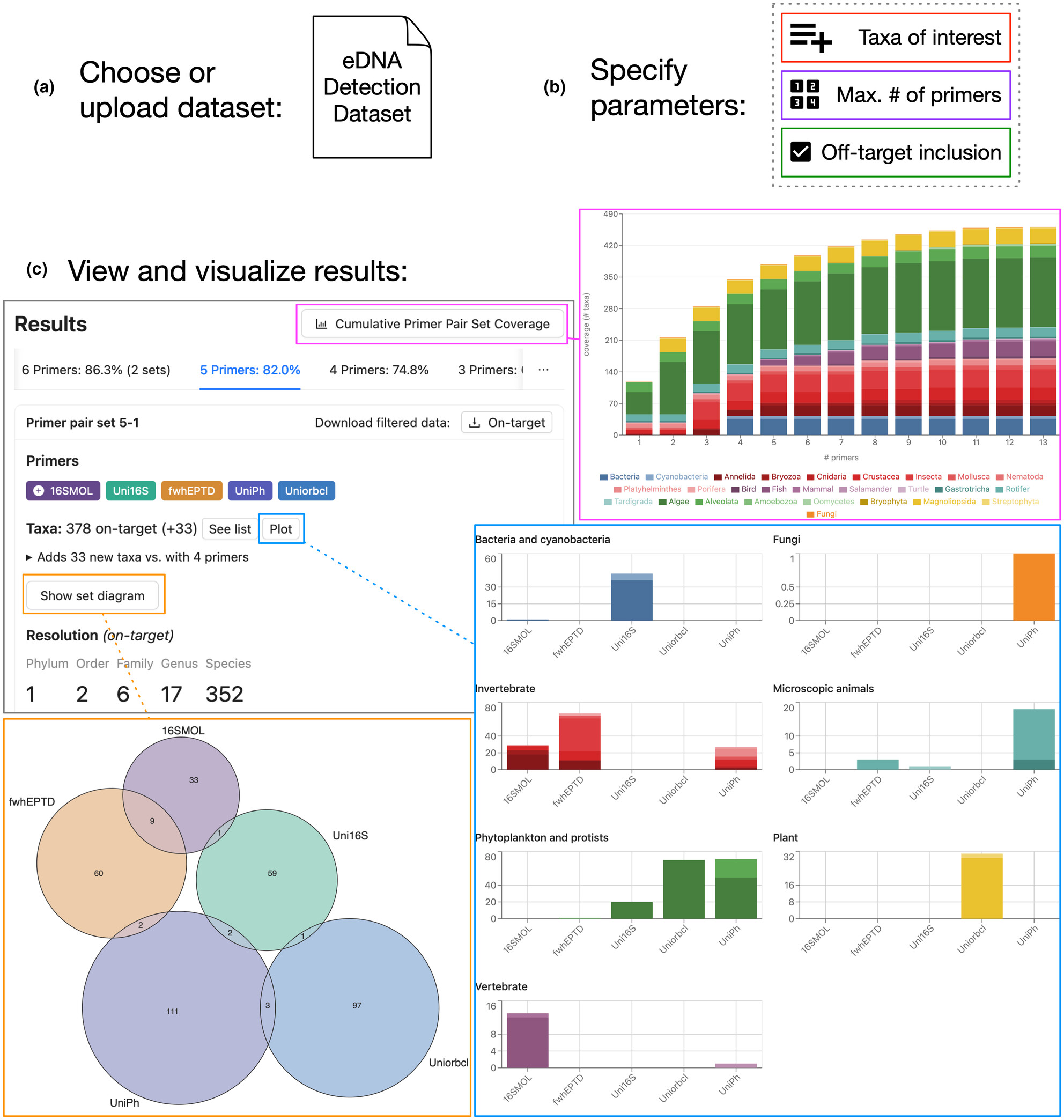
We used water samples from natural systems in Canada to create an eDNA metabarcoding dataset with 14 primer pairs that were evaluated in silico, in vitro (five mock communities), and in vivo (water samples from aquaria). The dataset was used to evaluate our new free, user-friendly online tool (SNIPe), created to derive subsets of informative, cost-effective primer pairs. We showed that 13 to 14 primer pairs recover 100% of the species in water samples (natural systems), but that four primer pairs are sufficient to recover almost 75% of taxa with little overlap. Our work highlights the power of eDNA metabarcoding for reconstructing freshwater communities, including prey, parasite, pathogen, invasive, and declining species. It also emphasizes the importance of marker choice on species resolution, as well as primer characteristics and filtering parameters on detection success and the accuracy of biodiversity estimates.
Environmental DNA reveals fine-scale spatial and temporal variation of marine mammals and their prey species in a Scottish marine protected area
- First Published: 22 July 2024
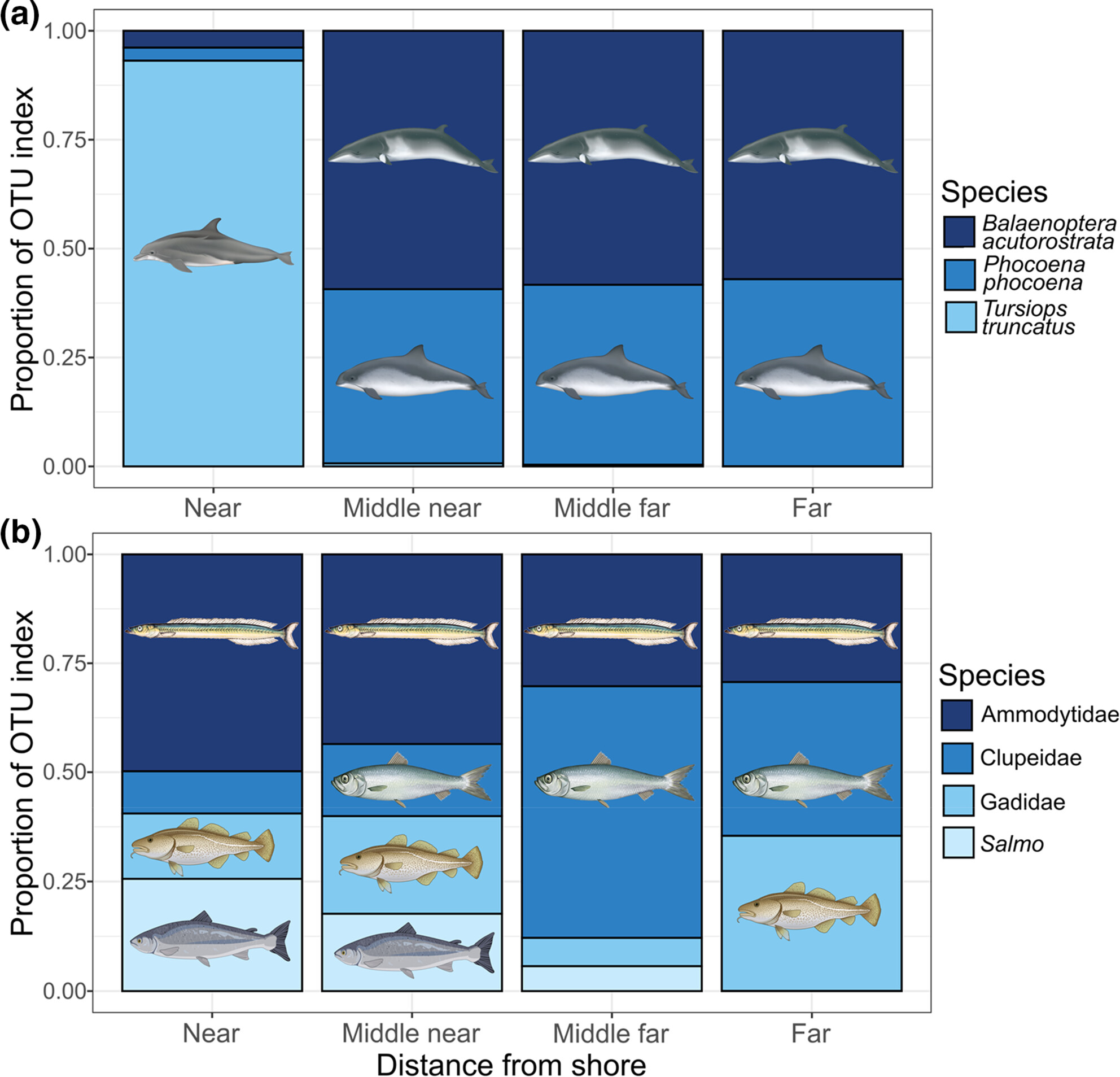
Prey are an important driver of fine-scale habitat use by marine mammals, but their distributions are often poorly understood. Here, we demonstrate that eDNA metabarcoding can reveal fine-scale spatiotemporal differences in cetacean distributions and their key prey species, including spatial partitioning between sympatric cetacean species with different dietary preferences and changing seasonal availability of key prey species.
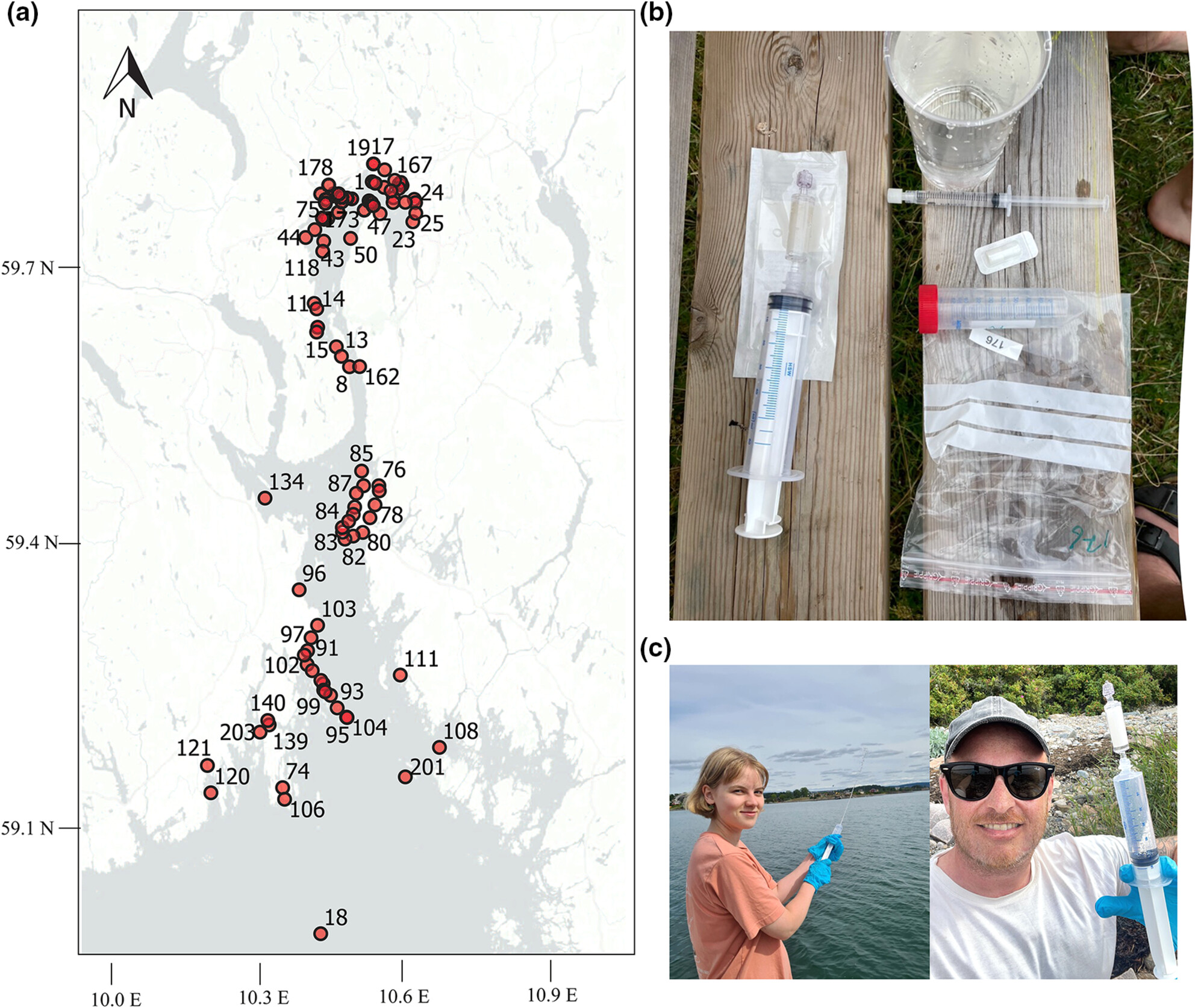
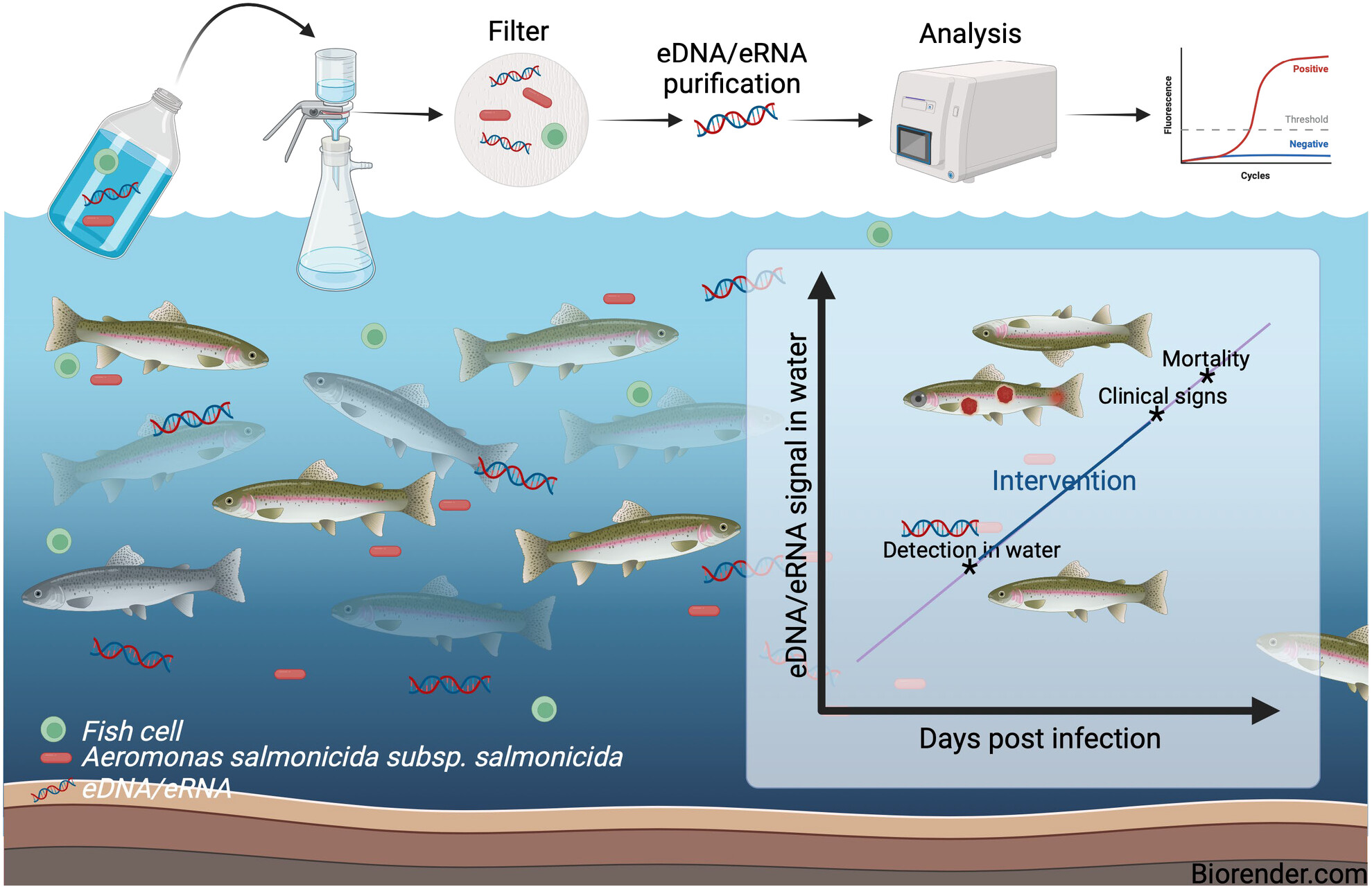
Investigation of the water environmental DNA/RNA profile for non-invasive biomonitoring of the fish pathogen Aeromonas salmonicida subsp. salmonicida and detection of immune responses in rainbow trout (Oncorhynchus mykiss)
- First Published: 24 July 2024
Enriching barcoding markers in environmental samples utilizing a phylogenetic probe design: Insights from mock communities
- First Published: 24 July 2024
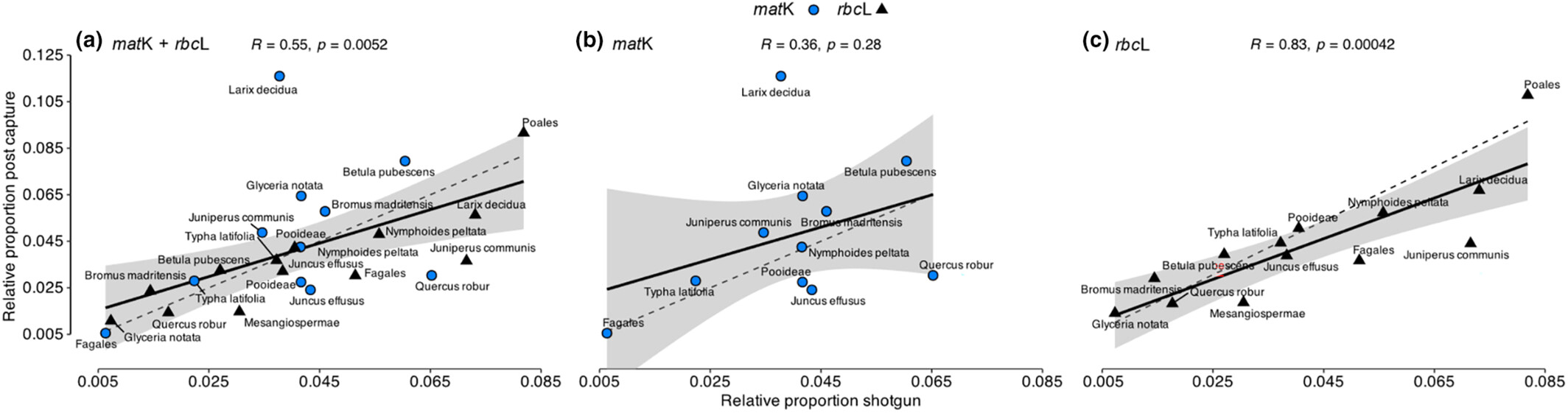
Hybridization capture to enrich environmental DNA libraries for specific organisms is offering an alternative to metabarcoding; however, little empirical work has addressed biases and implications for interpretations in molecular ecology. We used a phylogenetic-based approach to reduce the number of probes, assessed capture bias, and provided recommendations, based on insights gained from mock communities.
Disentangling eukaryotic biodiversity patterns from man-made environments (port and marina) and nearby coral reefs in the Red Sea: A focus on the surveillance of non-indigenous species
- First Published: 05 August 2024
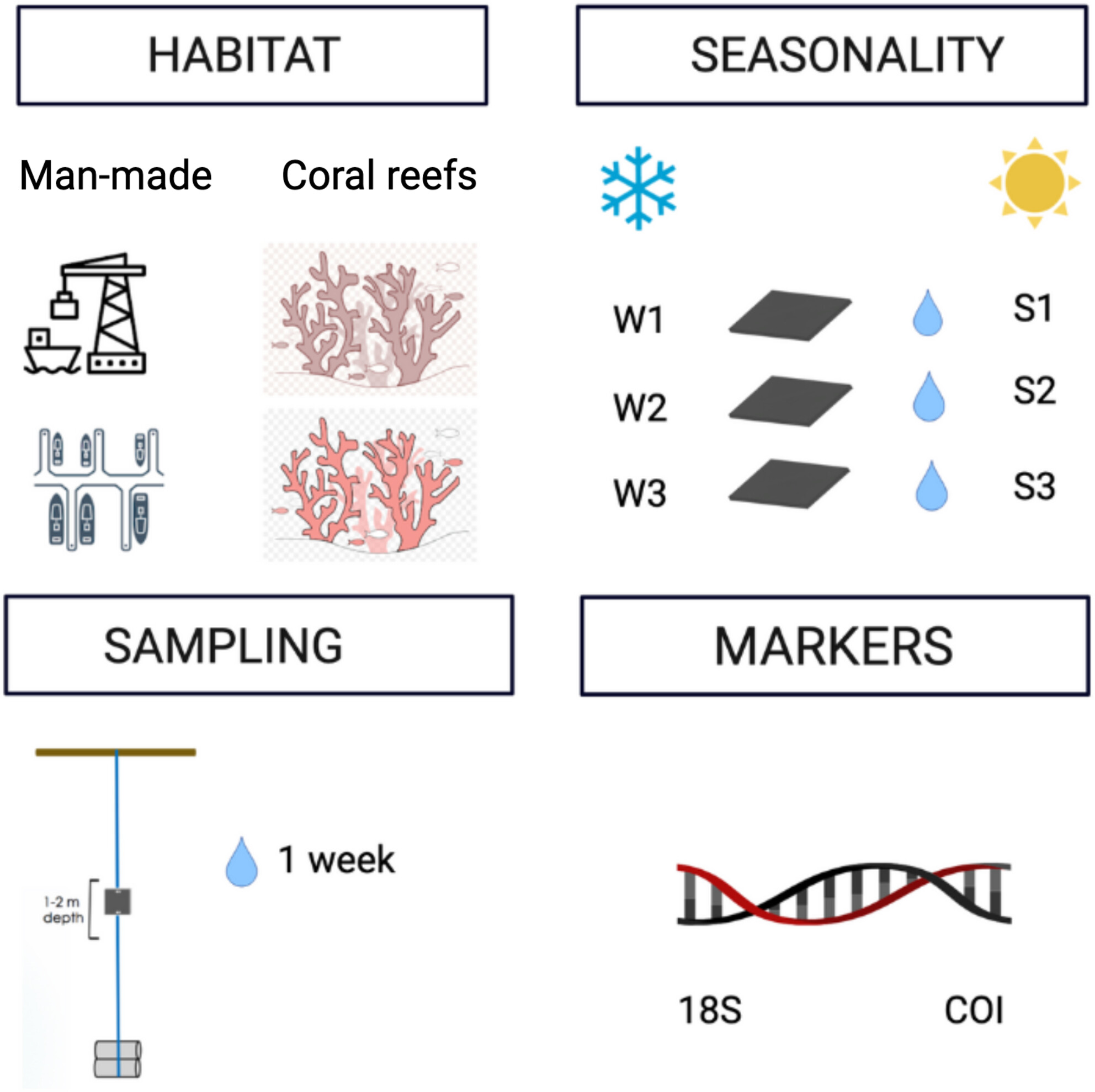
Our study focuses on the detection of invasive species across man-made environments and natural habitats in the central Red Sea. Specifically, we conducted comprehensive DNA analyses using samples collected from two artificial sites, a bustling port and a marina, in comparison with two ecologically sensitive natural sites, coral reefs. The aim was to assess the suitability of different sampling methods (PVC settlement panels and water samples) and genetic markers (18S rRNA and COI) to discern the presence of potential invasive species within these distinct environments.
Testing multiple environmental DNA substrates for detection of the cryptic and Critically Endangered burrowing freshwater crayfish Engaewa pseudoreducta
- First Published: 30 July 2024
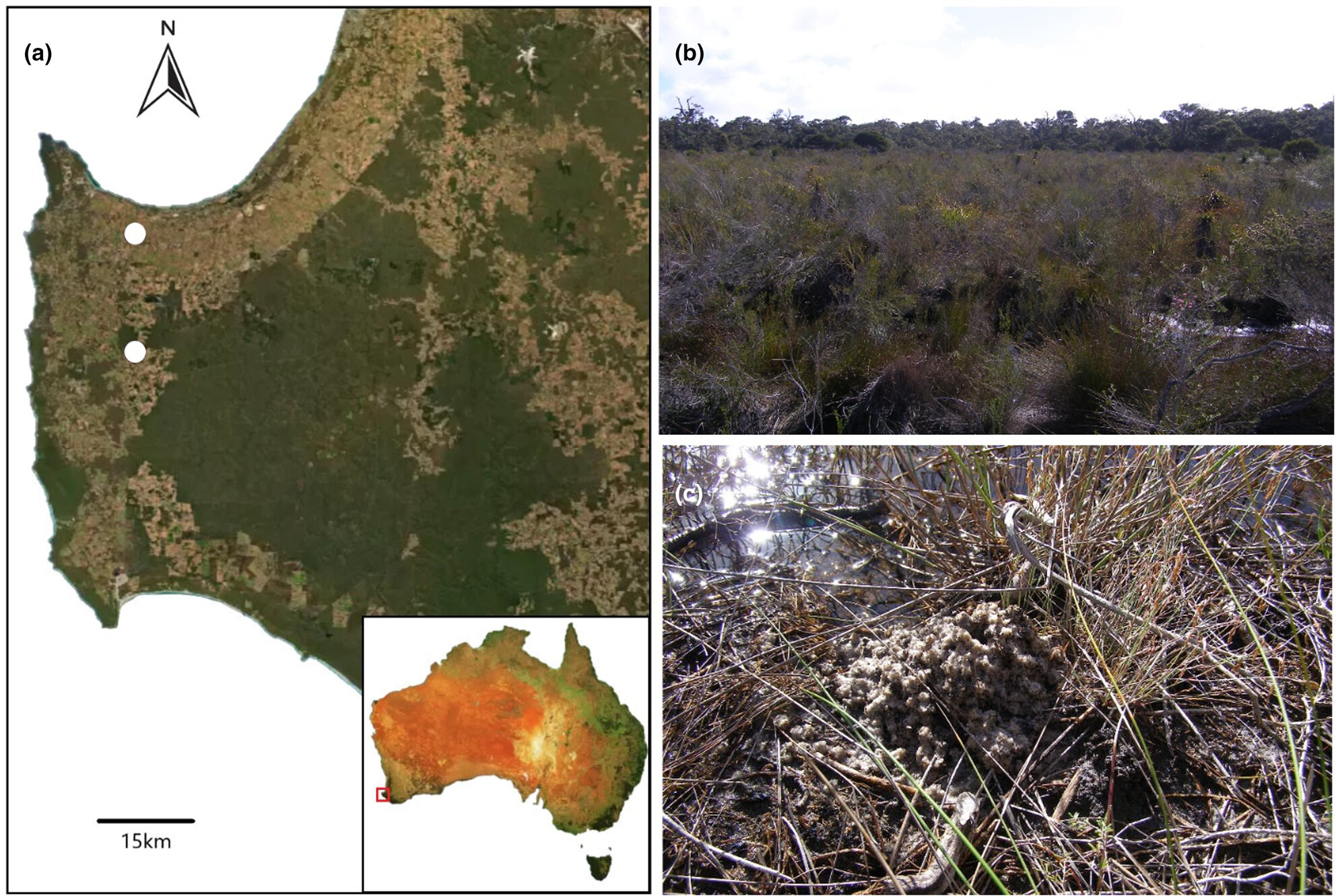
Conservation of subterranean species is challenging as this relies on distributional information, which is difficult to determine and often requires invasive sampling techniques. This study has designed a species-specific assay that can be used to test the environmental DNA that has been deposited on the surface by the burrowing crayfish Engaewa pseudoreducta. This assay has been validated through testing on multiple substrates and is the first study to our knowledge that has successfully applied this technique for a burrowing species.
Continuous daily sampling of airborne eDNA detects all vertebrate species identified by camera traps
- First Published: 22 August 2024
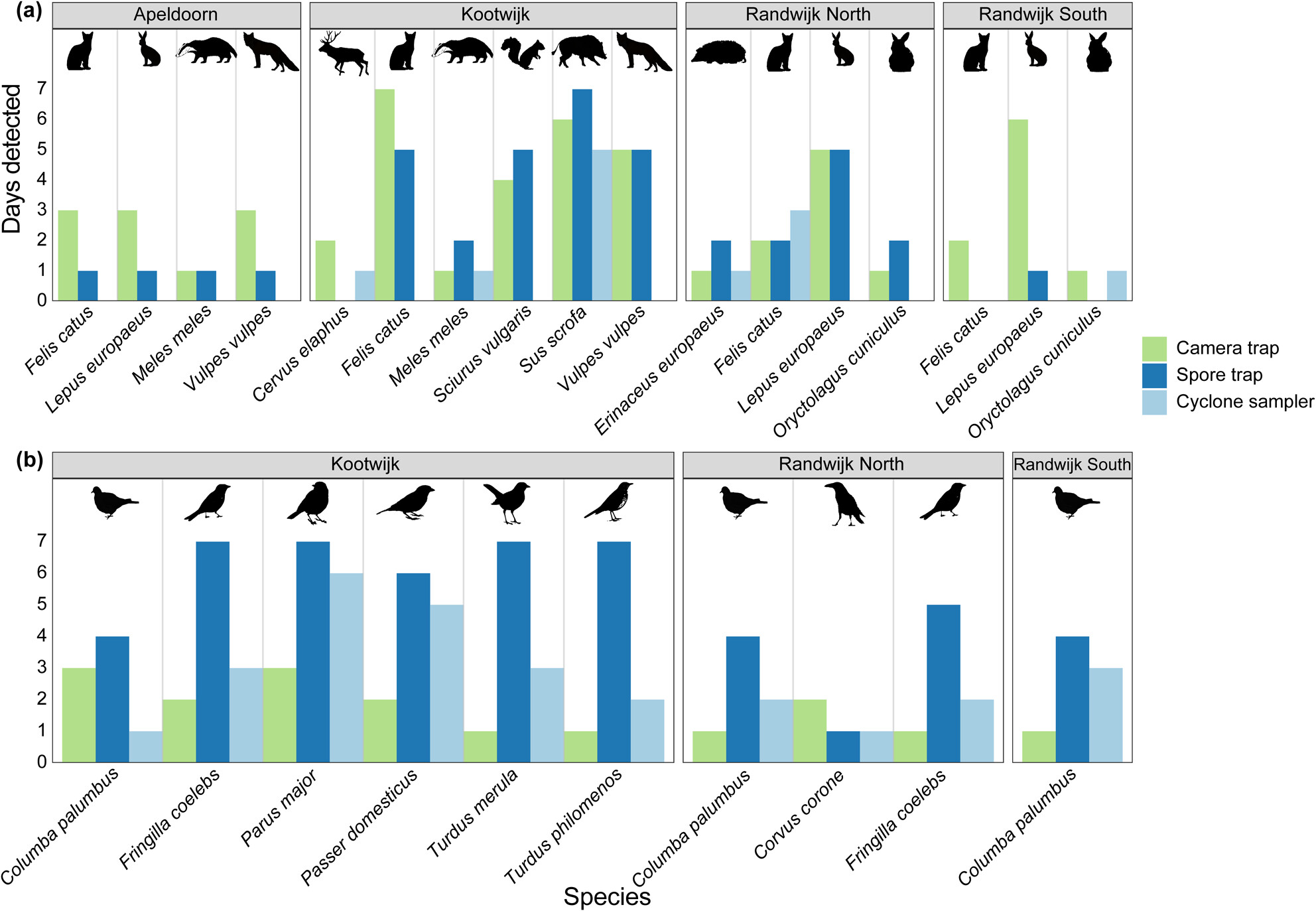
In this manuscript, we report on our study using airborne eDNA to monitor terrestrial vertebrate biodiversity in nature and compare the results with detections made using camera traps. Airborne eDNA detected all species identified using camera traps and many additional species. This indicates the sensitivity of the method, however, subsequent studies should prioritize validation of these findings through alternative biomonitoring approaches in order to gain better understanding of dispersal and fate of airborne eDNA.
Rapid and Portable Presumptive Loop-Mediated Isothermal Amplification Assays for the Detection of the Invasive Corn Snake (Pantherophis guttatus)
- First Published: 06 August 2024
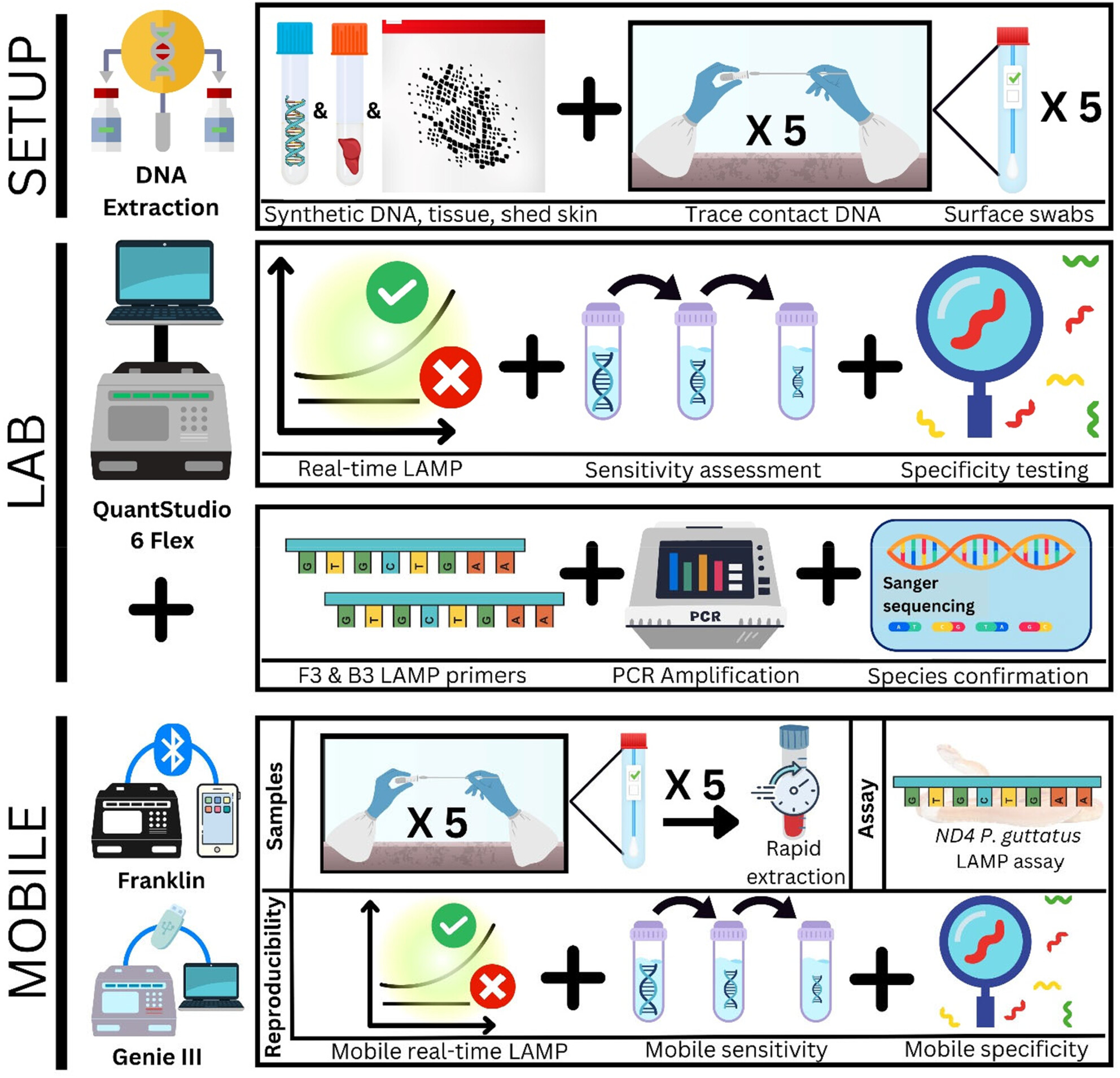
The international exotic pet trade is a major pathway for the introduction, establishment, and spread of novel invasive alien species. The North American corn snake (Pantherophis guttatus) is particularly common in the international pet trade and has been identified as a vertebrate pest priority species in Australia due to widespread climate suitability and prevalence in pre- and post-border seizure records. We developed two LAMP assays for the detection of P. guttatus, to provide tools suited to biosecurity detection for a species of highest vertebrate pest priority in Australia.
Comparison of Seven DNA Metabarcoding Sampling Methods to Assess Diet in a Large Avian Predator
- First Published: 20 August 2024
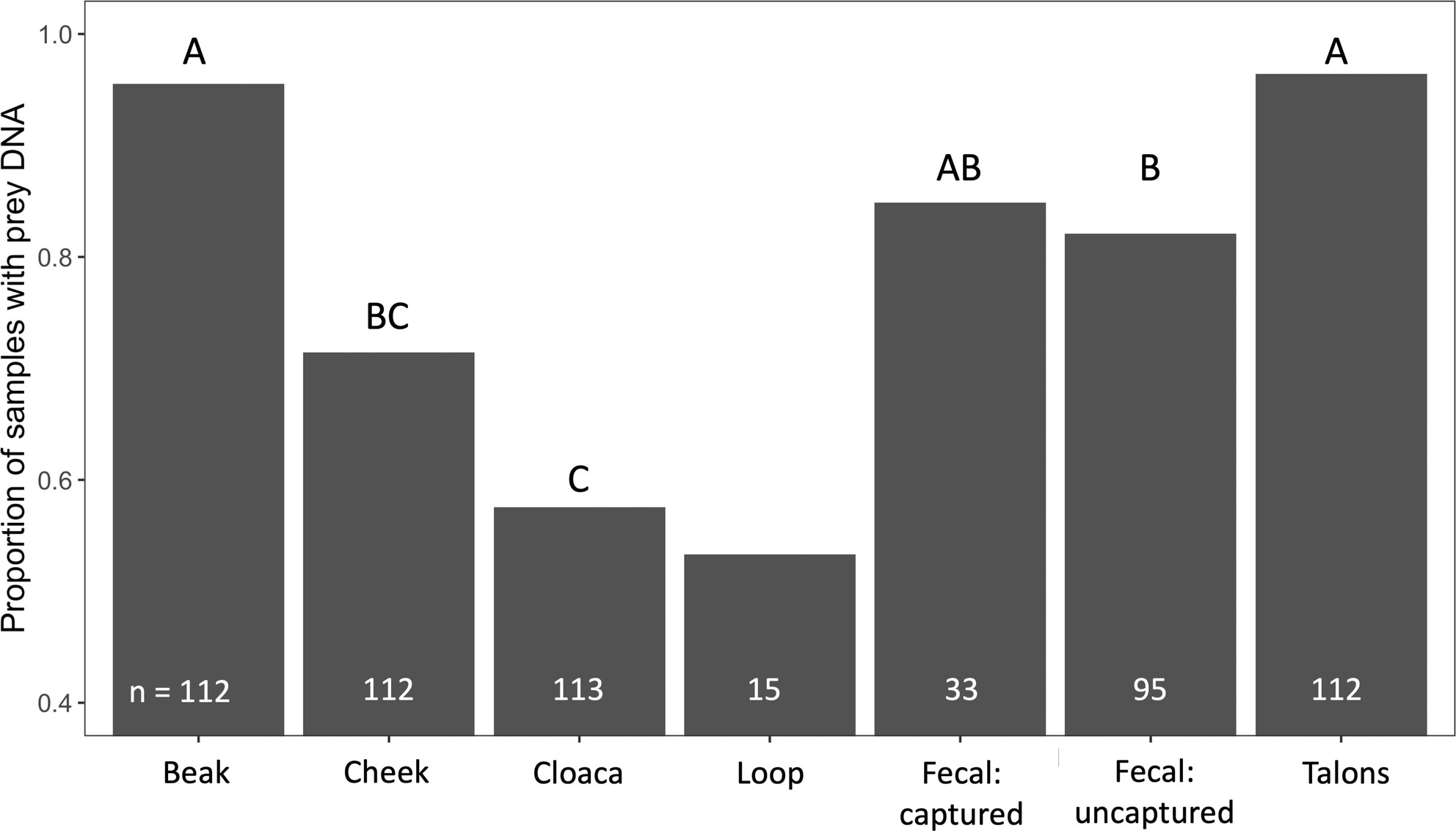
We evaluated seven DNA metabarcoding sampling methods to assess the diet of a large avian predator: Buteo lagopus (rough-legged hawk). While all sampling methods identified similar dominant prey taxa that were consistent with prior diet studies, swabbing the beak and talons produced greater prey detection rates and prey richness at both the individual and population levels.
eDNA Is a Useful Environmental Monitoring Tool for Assessing Stream Ecological Health
- First Published: 28 August 2024
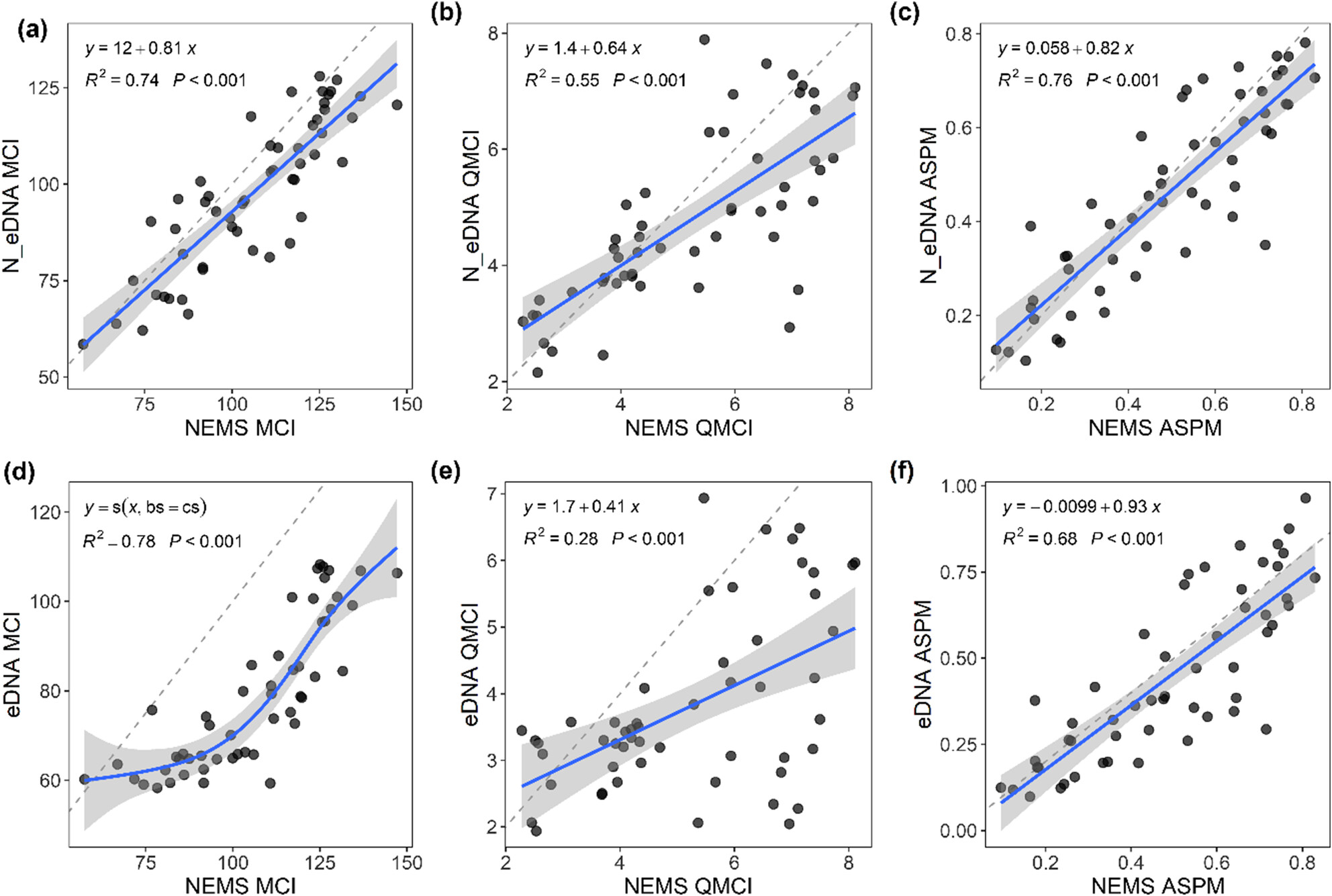
Ecosystem health assessments made by traditional biomonitoring techniques were compared to those made using eDNA at 53 sites throughout New Zealand. We found a high degree of correlation and convergence between biotic indices calculated from traditional biomonitoring techniques and from eDNA. Our results suggest that eDNA biomonitoring complements traditional methods and may be useful in meeting increased demands of future biomonitoring programs.