Journal list menu
Export Citations
Download PDFs
The Matchmaker Exchange: A Platform for Rare Disease Gene Discovery
- Pages: 915-921
- First Published: 13 August 2015
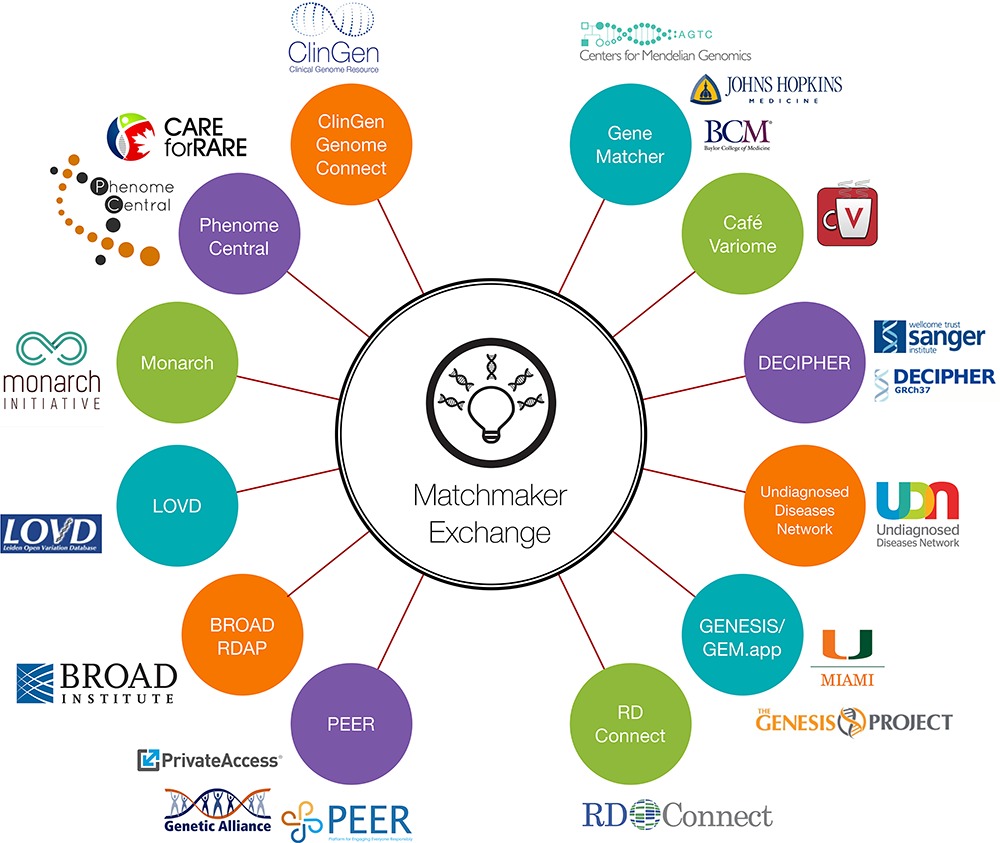
The Matchmaker Exchange (MME) includes representatives from the founding organizations and databases supporting or intending to support matchmaking services. Collaborative work has focused on both the technical aspects of data sharing, as well as policy considerations. This work has resulted in version 1.0 of a MME API, a set of requirements for qualifying as a MME service, and a user agreement for querying the MME.
The Matchmaker Exchange API: Automating Patient Matching Through the Exchange of Structured Phenotypic and Genotypic Profiles
- Pages: 922-927
- First Published: 08 August 2015
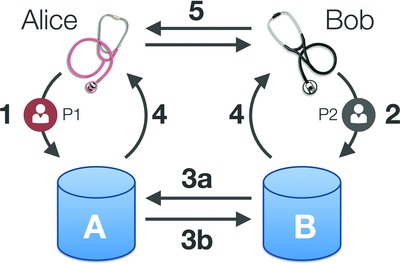
The Matchmaker Exchange API defines a protocol and data format for exchanging phenotype and genotype profiles between patient databases, in order to facilitate the identification of additional cohorts and increase the rate with which rare diseases can be researched and diagnosed. The API is straightforward and flexible in order to simplify its adoption on a large number of data types and workflows.
GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene
- Pages: 928-930
- First Published: 29 July 2015
PhenomeCentral: A Portal for Phenotypic and Genotypic Matchmaking of Patients with Rare Genetic Diseases
- Pages: 931-940
- First Published: 07 August 2015
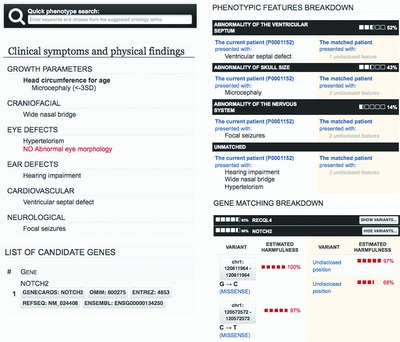
To enable the secure sharing of patient records between clinicians and rare disease scientists, we developed PhenomeCentral (https://phenomecentral.org), a restricted access portal for sharing de-identified patient phenotype and genotype data to find similar cases around the world. PhenomeCentral identifies similar patients based on semantic similarity between clinical features, automatically prioritized genes from whole-exome data, and candidate genes entered by the users, enabling both hypothesis-free and hypothesis-driven patient matchmaking.
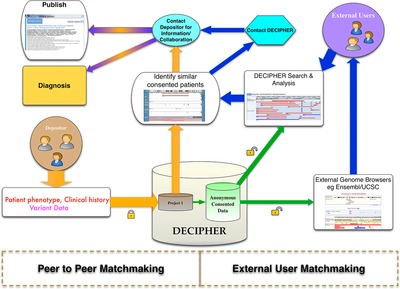
Facilitating Collaboration in Rare Genetic Disorders Through Effective Matchmaking in DECIPHER
- Pages: 941-949
- First Published: 29 July 2015
Innovative Genomic Collaboration Using the GENESIS (GEM.app) Platform
- Pages: 950-956
- First Published: 14 July 2015
Within GENESIS/GEM.app, our concept of user-driven real-time genomic data sharing and matchmaking is now the main engine for gene discoveries. In many of these findings, researchers from across the globe have been connected, which gave rise to the genetic evidence needed to successfully pinpoint specific gene mutations that explained patients' disease.
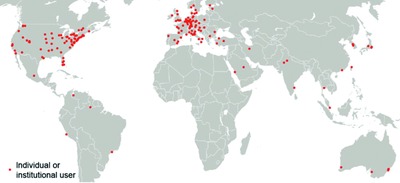
Cafe Variome: General-Purpose Software for Making Genotype–Phenotype Data Discoverable in Restricted or Open Access Contexts
- Pages: 957-964
- First Published: 29 July 2015
Data ‘discovery’ solutions enable users to quickly and accurately identify where useful data are located, thereby promoting collaboration around such datasets, and underpinning appropriate data sharing arrangements in restricted and open access contexts. Convenient, flexible, web-based, data discovery software is therefore needed, and Cafe Variome (www.cafevariome.org) represents one such tool that enables single groups or networks to advertise and/or share data in an appropriately controlled manner.
Participant-Driven Matchmaking in the Genomic Era
- Pages: 965-973
- First Published: 07 August 2015

Genomic matchmaking networks, which seek to link patients with similar genotypes and/or phenotypes, play an important role in confirming diagnoses made by whole-genome and -exome sequencing. We argue that matchmaking efforts led by affected individuals and their families provide a unique opportunity to greatly increase the size of these networks, and simultaneously increase the quantity and specificity of available data. We propose a participant-led technology solution to scale participant-led efforts, known as the Platform for Engaging Everyone Responsibly (PEER).
GenomeConnect: Matchmaking Between Patients, Clinical Laboratories, and Researchers to Improve Genomic Knowledge
- Pages: 974-978
- First Published: 16 July 2015

GenomeConnect is a project supported by the Clinical Genome Resource (ClinGen) providing the opportunity for patients to add to the genomics knowledge base by securely sharing health history and genetic test results. This paper covers a variety of topics including project development, the special roles of patients and laboratories, the relationship between GenomeConnect and the ClinVar database, the matchmaking capabilities of GenomeConnect, and the partnerships that GenomeConnect helps to form to increase data sharing and advance precision medicine.
Use of Model Organism and Disease Databases to Support Matchmaking for Human Disease Gene Discovery
- Pages: 979-984
- First Published: 13 August 2015
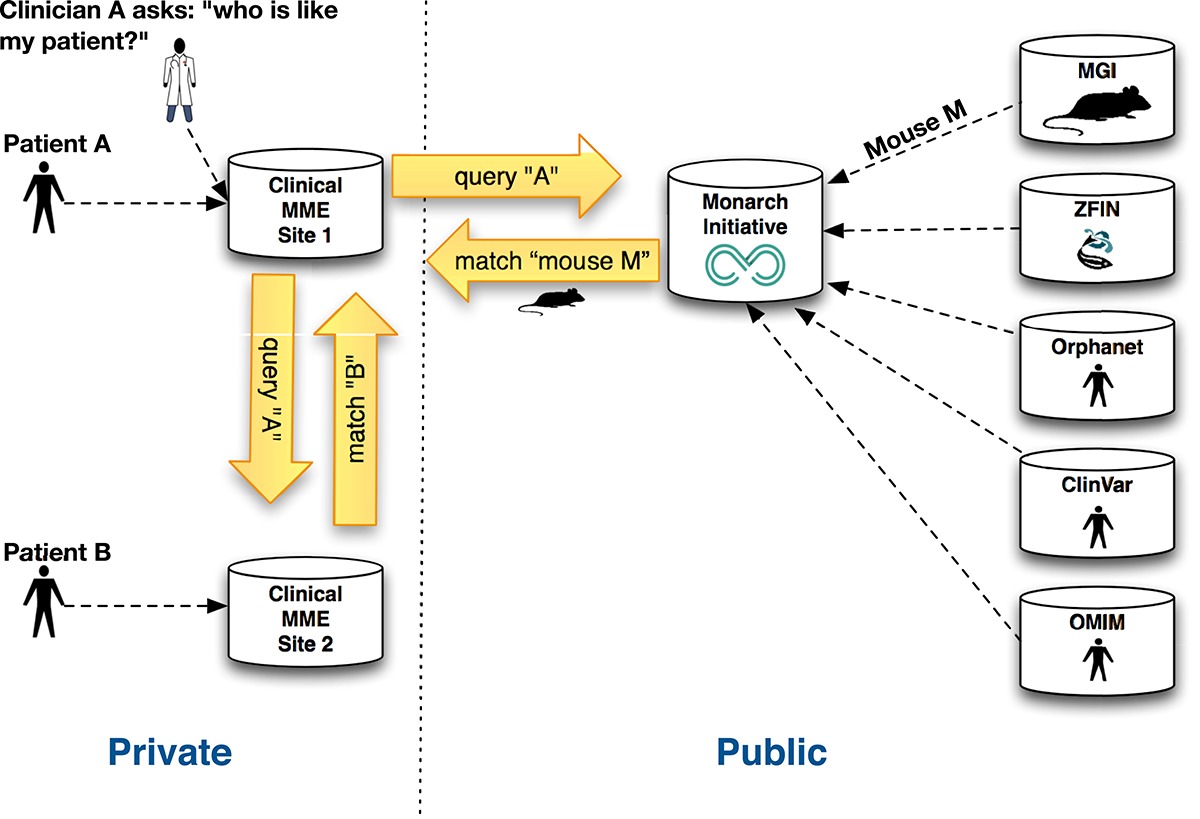
The Matchmaker exchange provides a system that allows cross-site phenotype search, to facilitate discovery of patients with matching variants or phenotypes. The Monarch Initiative expands the Matchmaker Exchange, providing a bridge to model organism genotype-phenotype resources.
Clinicians and researchers can broaden their search, looking not only for similar human patients, but for model systems that may be informative.
Data Sharing in the Undiagnosed Diseases Network
- Pages: 985-988
- First Published: 29 July 2015
The Genomic Birthday Paradox: How Much Is Enough?
- Pages: 989-997
- First Published: 02 August 2015
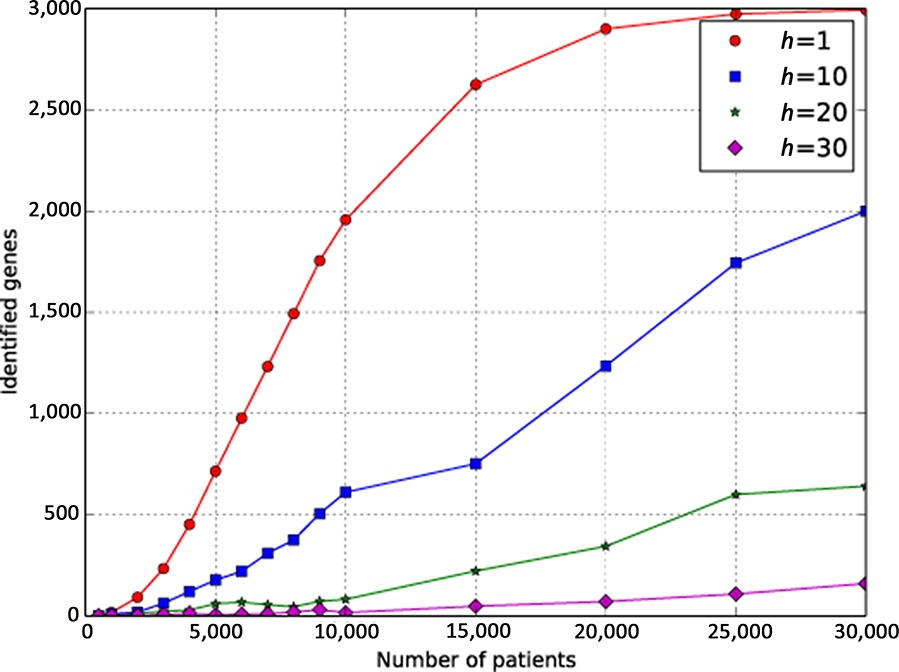
There are at least 3000 Mendelian disease-associated genes to be discovered. Genomic matchmaking databases (GMDs) allow participants to submit genotypic and phenotypic data in order to identify multiple comparable cases and thereby help to identify novel disease genes. Here, we present a series of simulations of GMDs to provide a an estimate of the number of cases that will be required.
Mitigating False-Positive Associations in Rare Disease Gene Discovery
- Pages: 998-1003
- First Published: 17 September 2015
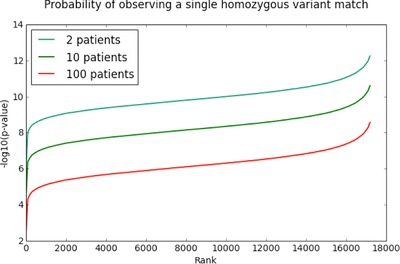
Unsolved clinical genetics cases can help identify new genes. However, by chance alone, it is often possible to find two individuals with the same phenotype that carry variants in the same gene. We quantify the risk of false-positive association in set of disease cases, using the prior probability of observing variants from over 60,000 individuals. Based on the number of individuals, cohort size, specific gene, and mode of inheritance, we calculate a p-value that the match represents a true association.
Novel COL2A1 Variant (c.619G>A, p.Gly207Arg) Manifesting as a Phenotype Similar to Progressive Pseudorheumatoid Dysplasia and Spondyloepiphyseal Dysplasia, Stanescu Type
- Pages: 1004-1008
- First Published: 17 July 2015
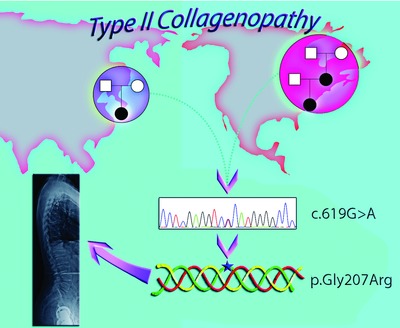
Here we report three individuals from two unrelated families who share the same novel COL2A1 variant and exhibit radiological features of progressive pseudorheumatoid dysplasia (PPRD) and Stanescu dysplasia. Whole exome sequencing in the first family identified a heterozygous COL2A1 variant (c.619G>A, p.Gly207Arg), which was independently detected by targeted COL2A1 sequencing in an unrelated proband with a similar phenotype. We suggest that the p.Gly207Arg variant causes a distinct type II collagenopathy with features of PPRD and Stanescu dysplasia.
GeneMatcher Aids in the Identification of a New Malformation Syndrome with Intellectual Disability, Unique Facial Dysmorphisms, and Skeletal and Connective Tissue Abnormalities Caused by De Novo Variants in HNRNPK
- Pages: 1009-1014
- First Published: 14 July 2015
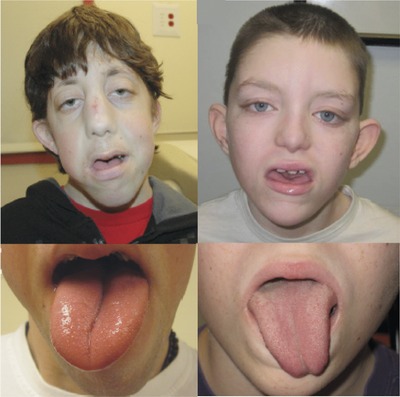
We report a new syndrome due to loss of function variants in the heterogeneous nuclear ribonucleoprotein K gene (HNRNPK, OMIM *600712). We describe two probands, both of whom have intellectual disability, a shared unique craniofacial phenotype, and connective tissue and skeletal abnormalities. The identification of this syndrome was made possible by a new online tool, GeneMatcher, which facilitates connections between clinicians and researchers based on shared interest in candidate genes, and highlights the newer paradigm of “reverse phenotyping.”
Matching Two Independent Cohorts Validates DPH1 as a Gene Responsible for Autosomal Recessive Intellectual Disability with Short Stature, Craniofacial, and Ectodermal Anomalies
- Pages: 1015-1019
- First Published: 29 July 2015
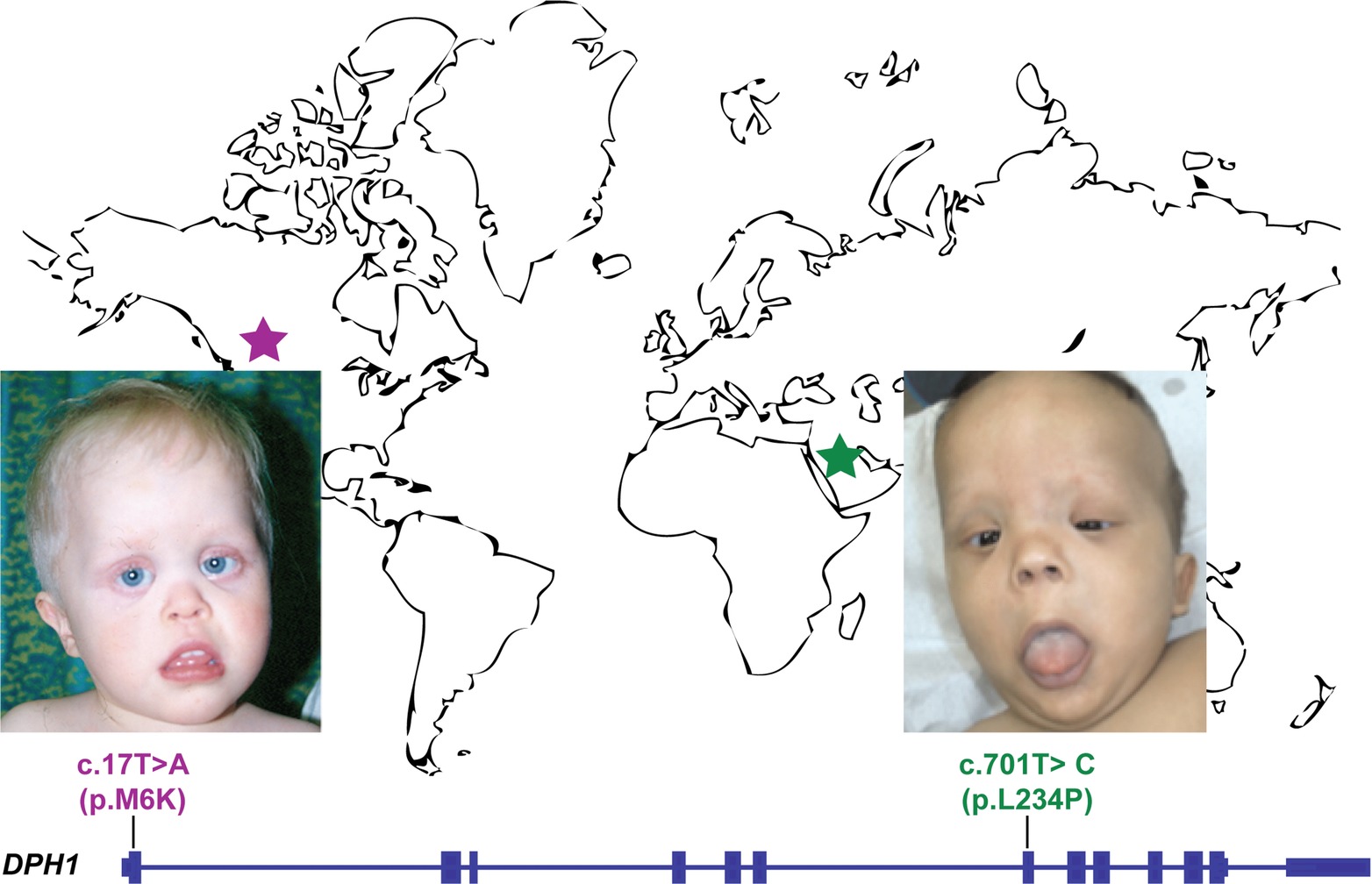
DPH1 was recently proposed as a candidate gene for syndromic intellectual disability after a homozygous mutation was associated with a 3C syndrome-like phenotype in four patients from a single extended family. Here we report a second homozygous missense variant in DPH1, seen in four members of a founder population, and associated with a phenotype initially reminiscent of Sensenbrenner syndrome. This post-publication ‘match’ validates DPH1 as a gene underlying syndromic intellectual disability with short stature and craniofacial and ectodermal anomalies, reminiscent of, but distinct from, 3C and Sensenbrenner syndromes. This validation took several years after the independent discoveries due to the absence of effective methods for sharing both candidate phenotype and genotype data between investigators, and underscores the value of novel mechanisms for matchmaking going forward.