Correlation between CTNNB1 mutation status and tumour phenotype in hepatitis B virus-related hepatocellular carcinoma
Yoon Jung Hwang
Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
Department of Pathology, Seoul National University Bundang Hospital, Seongnam, Korea
Search for more papers by this authorYangkyu Lee
Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
Department of Pathology, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
Search for more papers by this authorSu Jong Yu
Department of Internal Medicine and Liver Research Institute, Seoul National University College of Medicine; Biomedical Research Institute, Center for Medical Innovation, Seoul National University Hospital, Seoul, Korea
Search for more papers by this authorSuk Kyun Hong
Department of Surgery, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
Search for more papers by this authorNam-Joon Yi
Department of Surgery, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
Search for more papers by this authorYoungRok Choi
Department of Surgery, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
Search for more papers by this authorHyejung Lee
Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
Search for more papers by this authorWonju Chung
Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
Department of Pathology, Seoul National University Hospital, Seoul, Korea
Search for more papers by this authorCorresponding Author
Haeryoung Kim
Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
Department of Pathology, Seoul National University Hospital, Seoul, Korea
Address for correspondence: Haeryoung Kim, Department of Pathology, Seoul National University College of Medicine, 103 Daehak-no, Jongno-gu, Seoul 03080, Korea.
e-mail: [email protected]
Search for more papers by this authorYoon Jung Hwang
Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
Department of Pathology, Seoul National University Bundang Hospital, Seongnam, Korea
Search for more papers by this authorYangkyu Lee
Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
Department of Pathology, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
Search for more papers by this authorSu Jong Yu
Department of Internal Medicine and Liver Research Institute, Seoul National University College of Medicine; Biomedical Research Institute, Center for Medical Innovation, Seoul National University Hospital, Seoul, Korea
Search for more papers by this authorSuk Kyun Hong
Department of Surgery, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
Search for more papers by this authorNam-Joon Yi
Department of Surgery, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
Search for more papers by this authorYoungRok Choi
Department of Surgery, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
Search for more papers by this authorHyejung Lee
Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
Search for more papers by this authorWonju Chung
Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
Department of Pathology, Seoul National University Hospital, Seoul, Korea
Search for more papers by this authorCorresponding Author
Haeryoung Kim
Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
Department of Pathology, Seoul National University Hospital, Seoul, Korea
Address for correspondence: Haeryoung Kim, Department of Pathology, Seoul National University College of Medicine, 103 Daehak-no, Jongno-gu, Seoul 03080, Korea.
e-mail: [email protected]
Search for more papers by this authorAbstract
Aims
The frequency of CTNNB1 mutation, one of the most frequent genetic events in hepatocellular carcinoma (HCC), is lower in Asian countries and in hepatitis B virus (HBV)-related HCCs. In this study, we evaluated the prevalence and types of CTNNB1-mutation in HBV-related HCC and correlated the molecular status with the histomorphological and immunohistochemical features.
Methods and results
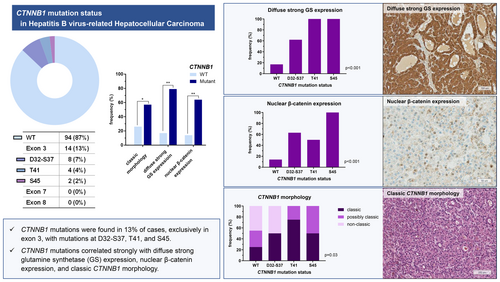
A total of 108 consecutive cases of treatment-naïve, surgically resected HBV-related HCCs were selected. Targeted sequencing for CTNNB1 exons 3, 7 and 8 was performed, and the results were correlated with the expression pattern of glutamine synthetase (GS), nuclear β-catenin expression status and the histomorphological characteristics of the tumour. CTNNB1 mutations were identified in 13% of HBV-related HCCs; of these cases, mutations were found in D32-S37 (7%), T41 (4%) and S45 (2%) of exon 3. None of the HCCs demonstrated alterations in exons 7 and 8. CTNNB1 mutation was strongly associated with diffuse strong GS expression (P < 0.001), nuclear β-catenin expression (P < 0.001) and the classic CTNNB1 morphology (P = 0.038). Diffuse strong GS expression was observed in 78.6% of the CTNNB1-mutated HCCs, and nuclear β-catenin expression was identified in 64.3% of these cases. The classic CTNNB1 morphology was observed in 57% of all CTNNB1-mutated HCCs. Furthermore, programmed death-ligand 1 (PD-L1) was less frequently expressed in HCCs with classic CTNNB1 morphology.
Conclusions
CTNNB1 mutation was observed in 13% of HBV-related HCCs in this Korean cohort, and was associated with diffuse strong GS expression, nuclear β-catenin expression and classic CTNNB1 morphology.
Graphical Abstract
Open Research
Data availability
All data generated or analysed during this study are included in this article and its supporting information files. Further enquiries can be directed to the corresponding author.
Supporting Information
| Filename | Description |
|---|---|
| his15363-sup-0001-TableS1.zipZip archive, 66.8 KB |
Table S1: The sequence of primers used for amplification of CTNNB1 gene. Table S2: List of CTNNB1-mutated HBV-related HCC cases. Table S3: Comparison of clinicopathologic characteristics according to CTNNB1 mutation status. Table S4: Comparison of clinicopathologic characteristics according to diffuse strong glutamine synthetase expression. Table S5: Comparison of clinicopathologic characteristics according to nuclear β-catenin expression. Table S6: Comparison of clinicopathologic characteristics according to CTNNB1 morphology. Table S7: Clinicopathological details of HBV-related HCC cases with CTNNB1 mutation (next-generation sequencing cohort). |
| his15363-sup-0002-FigureS1.jpgJPEG image, 106.6 KB |
Figure S1: Recurrence-free survival of hepatocellular carcinoma patients, stratified by CTNNB1 mutation status (A), glutamine synthetase (GS) expression pattern (B), nuclear β-catenin expression status (C) and CTNNB1 morphology (D). |
| his15363-sup-0003-FigureS2.tifTIFF image, 226.6 KB |
Figure S2: CTNNB1 mutation status in HBV-related HCC cases of the next-generation sequencing (NGS) cohort (A, n = 24) and the total cohort (B, n = 132). Frequency of CTNNB1 morphology according to CTNNB1 mutation status (C). Frequency of histological features according to CTNNB1 mutation status (D) (MTM: macrotrabecular massive subtype; VETC: vessels encapsulating tumour clusters pattern). |
| his15363-sup-0004-FigureS3.tifTIFF image, 46.6 KB |
Figure S3: Differences in tumour size (A) and frequencies of macrotrabecular massive (MTM) subtype and vessels encapsulating tumour clusters (VETC) pattern (B) according to the presence of tumour necrosis. |
| his15363-sup-0005-FigureS4.tifTIFF image, 27.5 KB |
Figure S4: Vessels encapsulating tumour clusters (VETC) pattern and macrotrabecular-massive (MTM) subtype according to glutamine synthetase (GS) expression pattern in tissue microarray cohort. |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
REFERENCES
- 1 Cancer Genome Atlas Research Network. Electronic address wbe, cancer genome atlas research N. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017; 169; e1323.
- 2Llovet JM, Kelley RK, Villanueva A et al. Hepatocellular carcinoma. Nat Rev Dis Primers 2021; 7; 6.
- 3Torbenson M, McCabe CE, O'Brien DR et al. Morphological heterogeneity in beta-catenin-mutated hepatocellular carcinomas: Implications for tumor molecular classification. Hum Pathol 2022; 119; 15–27.
- 4Zucman-Rossi J, Villanueva A, Nault JC, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology 2015; 149; 1226–1239. e4.
- 5Calderaro J, Ziol M, Paradis V, Zucman-Rossi J. Molecular and histological correlations in liver cancer. J Hepatol 2019; 71; 616–630.
- 6Guichard C, Amaddeo G, Imbeaud S et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet 2012; 44; 694–698.
- 7Austinat M, Dunsch R, Wittekind C, Tannapfel A, Gebhardt R, Gaunitz F. Correlation between beta-catenin mutations and expression of wnt-signaling target genes in hepatocellular carcinoma. Mol Cancer 2008; 7; 21.
- 8MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: Components, mechanisms, and diseases. Dev Cell 2009; 17; 9–26.
- 9Xu C, Xu Z, Zhang Y, Evert M, Calvisi DF, Chen X. Beta-catenin signaling in hepatocellular carcinoma. J Clin Invest 2022; 132; e154515.
- 10Joseph NM, Umetsu SE, Shafizadeh N, Ferrell L, Kakar S. Genomic profiling of well-differentiated hepatocellular neoplasms with diffuse glutamine synthetase staining reveals similar genetics across the adenoma to carcinoma spectrum. Mod Pathol 2019; 32; 1627–1636.
- 11Rebouissou S, Franconi A, Calderaro J et al. Genotype-phenotype correlation of ctnnb1 mutations reveals different ss-catenin activity associated with liver tumor progression. Hepatology 2016; 64; 2047–2061.
- 12Hale G, Liu X, Hu J et al. Correlation of exon 3 beta-catenin mutations with glutamine synthetase staining patterns in hepatocellular adenoma and hepatocellular carcinoma. Mod Pathol 2016; 29; 1370–1380.
- 13Joseph NM, Ferrell LD, Jain D et al. Diagnostic utility and limitations of glutamine synthetase and serum amyloid-associated protein immunohistochemistry in the distinction of focal nodular hyperplasia and inflammatory hepatocellular adenoma. Mod Pathol 2014; 27; 62–72.
- 14Cieply B, Zeng G, Proverbs-Singh T, Geller DA, Monga SP. Unique phenotype of hepatocellular cancers with exon-3 mutations in beta-catenin gene. Hepatology 2009; 49; 821–831.
- 15Calderaro J, Couchy G, Imbeaud S et al. Histological subtypes of hepatocellular carcinoma are related to gene mutations and molecular tumour classification. J Hepatol 2017; 67; 727–738.
- 16Tornesello ML, Buonaguro L, Tatangelo F, Botti G, Izzo F, Buonaguro FM. Mutations in tp53, ctnnb1 and pik3ca genes in hepatocellular carcinoma associated with hepatitis b and hepatitis c virus infections. Genomics 2013; 102; 74–83.
- 17Pezzuto F, Izzo F, Buonaguro L et al. Tumor specific mutations in tert promoter and ctnnb1 gene in hepatitis b and hepatitis c related hepatocellular carcinoma. Oncotarget 2016; 7; 54253–54262.
- 18Khalaf AM, Fuentes D, Morshid AI et al. Role of wnt/beta-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. J Hepatocell Carcinoma 2018; 5; 61–73.
- 19Wang W, Pan Q, Fuhler GM, Smits R, Peppelenbosch MP. Action and function of wnt/beta-catenin signaling in the progression from chronic hepatitis c to hepatocellular carcinoma. J Gastroenterol 2017; 52; 419–431.
- 20Javanmard D, Najafi M, Babaei MR et al. Investigation of ctnnb1 gene mutations and expression in hepatocellular carcinoma and cirrhosis in association with hepatitis b virus infection. Infect Agent Cancer 2020; 15; 37.
- 21Liu K, Dennis C, Prince DS et al. Vessels that encapsulate tumour clusters vascular pattern in hepatocellular carcinoma. JHEP Rep 2023; 5; 100792.
- 22Hendry S, Salgado R, Gevaert T et al. Assessing tumor-infiltrating lymphocytes in solid tumors: A practical review for pathologists and proposal for a standardized method from the international immuno-oncology biomarkers working group: Part 2: Tils in melanoma, gastrointestinal tract carcinomas, non-small cell lung carcinoma and mesothelioma, endometrial and ovarian carcinomas, squamous cell carcinoma of the head and neck, genitourinary carcinomas, and primary brain tumors. Adv Anat Pathol 2017; 24; 311–335.
- 23Ahn SM, Jang SJ, Shim JH et al. Genomic portrait of resectable hepatocellular carcinomas: Implications of rb1 and fgf19 aberrations for patient stratification. Hepatology 2014; 60; 1972–1982.
- 24Kim S, Jeong S. Mutation hotspots in the beta-catenin gene: Lessons from the human cancer genome databases. Mol Cells 2019; 42; 8–16.
- 25Lee SE, Chang SH, Kim WY et al. Frequent somatic tert promoter mutations and ctnnb1 mutations in hepatocellular carcinoma. Oncotarget 2016; 7; 69267–69275.
- 26Huang SC, Koch CA, Vortmeyer AO et al. Duplication of the mutant ret allele in trisomy 10 or loss of the wild-type allele in multiple endocrine neoplasia type 2-associated pheochromocytomas. Cancer Res 2000; 60; 6223–6226.
- 27Graveel C, Su Y, Koeman J et al. Activating met mutations produce unique tumor profiles in mice with selective duplication of the mutant allele. Proc Natl Acad Sci USA 2004; 101; 17198–17203.
- 28Kralovics R, Passamonti F, Buser AS et al. A gain-of-function mutation of jak2 in myeloproliferative disorders. N Engl J Med 2005; 352; 1779–1790.
- 29Fitzgibbon J, Smith LL, Raghavan M et al. Association between acquired uniparental disomy and homozygous gene mutation in acute myeloid leukemias. Cancer Res 2005; 65; 9152–9154.
- 30Hsieh A, Kim HS, Lim SO, Yu DY, Jung G. Hepatitis b viral x protein interacts with tumor suppressor adenomatous polyposis coli to activate wnt/beta-catenin signaling. Cancer Lett 2011; 300; 162–172.
- 31Harding JJ, Nandakumar S, Armenia J et al. Prospective genotyping of hepatocellular carcinoma: Clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin Cancer Res 2019; 25; 2116–2126.
- 32Aoki T, Nishida N, Kurebayashi Y et al. Two distinct characteristics of immune microenvironment in human hepatocellular carcinoma with wnt/beta-catenin mutations. Liver Cancer 2024; 13; 285–305.
- 33Renne SL, Woo HY, Allegra S et al. Vessels encapsulating tumor clusters (vetc) is a powerful predictor of aggressive hepatocellular carcinoma. Hepatology 2020; 71; 183–195.
- 34Kurebayashi Y, Matsuda K, Ueno A et al. Immunovascular classification of hcc reflects reciprocal interaction between immune and angiogenic tumor microenvironments. Hepatology 2022; 75; 1139–1153.
- 35Matsuda K, Kurebayashi Y, Masugi Y et al. Immunovascular microenvironment in relation to prognostic heterogeneity of wnt/beta-catenin-activated hepatocellular carcinoma. Hepatol Res 2023; 53; 344–356.
- 36Fang JH, Zhou HC, Zhang C et al. A novel vascular pattern promotes metastasis of hepatocellular carcinoma in an epithelial-mesenchymal transition-independent manner. Hepatology 2015; 62; 452–465.
- 37Fujii T, Kuwano H. Regulation of the expression balance of angiopoietin-1 and angiopoietin-2 by shh and fgf-2. In Vitro Cell Dev Biol Anim 2010; 46; 487–491.
- 38Rebouissou S, Nault JC. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J Hepatol 2020; 72; 215–229.