Liquid biopsy: A new avenue in pathology
Abstract
Liquid biopsy is a relatively new entity. This non-invasive technique provides real-time information about a tumour. The liquid biopsy contains circulating tumour cells, cell-free DNA and exosomes. The main indications for liquid biopsy include early diagnosis, screening, detection of minimal residual disease, designing personalised treatment and predicting biological behaviour of the tumour. In this review, we discuss various aspects of liquid biopsy and compare it with conventional biopsy.
Abstract
In this article, the various aspects of liquid biopsy were discussed and compared with the conventional biopsy.
1 INTRODUCTION
The term liquid biopsy is a relatively recent introduction in the field of tumour pathology. It is a new concept in the diagnosis, management and predictive behaviour of a tumour. Liquid biopsy was originally defined by National Cancer Institute as “the test on the blood sample of the patient to study the circulating tumour cells or cell free DNA derived from the tumour cells in the blood.”1 In recent years, liquid biopsy has played an important role for real-time monitoring of the various aspects of the tumour. It is one of the most promising technologies in the field of tumour pathology. In this review, we discuss the various aspects of liquid biopsy including the detection, analysis, clinical applications and its future prospects.
1.1 The components of liquid biopsy
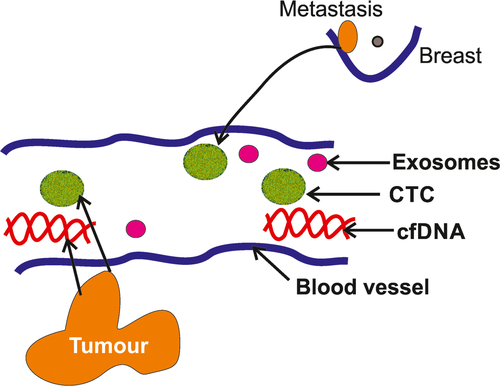
The conventional biopsy contains cells or tissues from the lesion. However, the liquid biopsy includes circulating tumour cells (CTCs), cell-free DNA (cfDNA) and exosomes2, 3 (Figure 1). During the initial period of oncogenesis and metastasis, the primary tumours shed the whole cell, cfDNA or exosomes (containing mRNA, micro- [mi]-RNA or small RNA [s]RNA) that carry vital information of the primary tumour.
2 BIOLOGY OF THE LIQUID BIOPSY
2.1 CTCs
The sources of the CTCs are both from the primary (mainly) and metastatic sites. The certain clone of the primary tumour comes to the circulation and disseminates to the distant organs. These CTCs may be actively released from the main tumour or they may be just the apoptotic cell. The overall half-life of CTCs is only 1-2 hours. Subsequently, the CTCs are removed from the circulation either by clearance in the liver or by infiltration in the metastatic sites. The circulating tumour cells can be identified by the presence of cytokeratin and epithelial cell adhesion molecule (EpCAM) on their cell surface along with the absence of CD 45.
2.2 cfDNA
The sources of cfDNA of tumour cells in the blood circulation include: (a) the tumour proper; (b) CTCs; and (c) metastatic tumour. cfDNA is derived from the apoptotic cell, necrotic tumour cells, active release from the viable tumour cells and also from the destruction of CTCs. cfDNA at first comes into circulation as a nucleosomal complex that contains both DNA and histones and subsequently DNA is released from the nucleosome complex. These cfDNAs are double-stranded and have much lower molecular weight than that of genomic DNA because of fragmentation in the circulation. The half-life of cfDNA is 16-30 minutes and it is possibly cleared mainly by liver and kidney.4, 5
2.3 Extracellular vesicles
Extracellular vesicles are another component of liquid biopsy that are released both from the normal as well as tumour cells and are found in the both blood and in the various body fluids. They are membrane-bound vesicles and consist of exosomes, microvesicles and apoptotic bodies. The exosomes are nano-sized vesicles whereas microvesicles are micron-sized vesicles and larger in size. The exosomes contain protein, lipids, mRNA, DNA, mi-RNA and sRNA and carry many tumour-specific valuable information.6 They are produced within the endoplasmic networks and are released from the plasma membrane of the tumour cells. Subsequently, the exosomes are removed from the circulation with the help of phagocytosis of the recipient cells. The half-life of the exosomes is short and 90% are cleared from the circulation within 30 minutes.7
3 COLLECTION AND ENRICHMENT OF THE CONTENTS OF LIQUID BIOPSY
3.1 CTCs
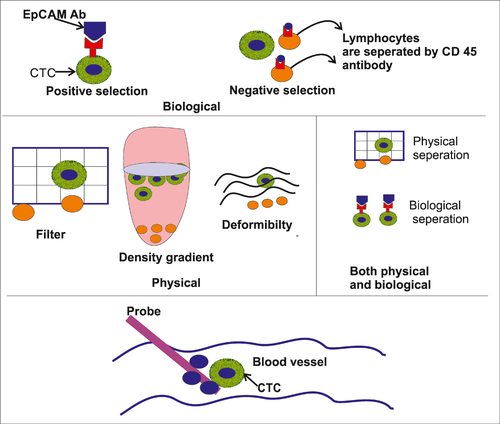
The detection, isolation and enrichment of CTCs are the great technological challenge as there are too few in the circulation, with approximately one CTC present per 100 million cells in the blood (Figure 2). Therefore, extremely sensitive and specific technology is needed for the detection and separation of CTS. The following strategies may be taken for CTC enrichment:
3.1.1 Biological properties
- Immunological assays: Here, antibodies against cell surface antigens are used to separate the tumour cells. Positive selection: In case of positive selection, the antibodies are tagged with magnetic bead and subsequently the cells are exposed to a magnetic field to separate the antibody-tagged cells. Antibodies against EpCAM or a combination of anti-CK and anti-EpCAM antibodies are used in positive selection. The CTCs are further confirmed by using a panel of antibodies consisting of CK 8, CK 18, CK 19, CD 45 and DAPI. CellSearch system uses the immunomagnetic beads to detect and separate the CTCs. This is the only Food and Drug Administration-approved system. Negative selection: In negative selection, the lymphoid cells are separated by using anti-CD 45 antibodies.
- Microfluidic chips: Here, immunomagnetic based microfluidic chips are used to capture CTCs and this system can separate the cells from a very small volume of sample.
3.1.2 Physical properties
The various physical properties of CTCs can be used for detection and separation of the cells. Size, a variation of deformability, density and surface electrical charges are mainly utilised to detect and separate CTCs.
3.1.3 Both physical and immunological properties
Immunomagnetic separation of the tumour cells along with the manipulation of physical properties of CTCs can be used.
3.1.4 Others
Recently, a novel technique has been used for enrichment of the CTCs in vivo by inserting a nanodetector in the arm vein of the patient. The detector can pick up CTCs from at least 1.5 L of flowing blood and therefore the chance of getting the tumour cells is high due to the large volume of blood is sampled in vivo.
3.2 Cell-free DNA
Cell-free DNA can be removed from the blood by simple phenol/chloroform extraction technique. It also can be extracted by silica-gel membrane technique, fluorometric technique using various dyes, such as SYBER Green I, and polymerase chain reaction (PCR) assays. The fluorometric assay provides an estimation of the total cfDNA content, whereas PCR-based assay helps in the detection of the very small DNA sequence of tumour-derived DNA.
3.3 Exosomes
Exosomes are found in blood, and also in various body fluids such as urine, CSF, effusion fluid and saliva. The exosomes can be isolated from the body fluid and stored for many years.8 The exosomes are usually abundant in the blood and they bear the surface markers of the original cells. The exosomes can be extracted with the help of antibody-based arrays, immunoaffinity-based capture technique and microfluidic chip methods.9
4 ANALYSIS OF LIQUID BIOPSY
The liquid biopsy contents can be analysed for the assessment of various changes (Table 1).
| Factors to assess | Techniques applied | Performed on |
|---|---|---|
| Mutational detection | Amplification-refractory mutation system, BEAMing, ddPCR, ice-COLD PCR and next-generation sequencing | CTCs and cell-free DNA |
| Epigenetic study | Methylation-specific polymerase chain reaction protocol, microarray-based DNA methylation assay | CTCs and cell-free DNA |
| Study of RNA (micro RNA and small RNA) | Quantitative reverse transcriptase PCR | Exosomes |
| Tumour cell phenotyping | Immunocytochemistry | CTCs |
- CTC, circulating tumour cell; PCR, polymerase chain reaction.
4.1 Mutational detection
The detection of mutation in liquid biopsy is the most important and difficult task. Various techniques can be used for this purpose, including amplification-refractory mutation system (ARMS), BEAMing, droplet digital (dd)PCR, ice-cold PCR and next-generation sequencing (NGS).10-12
BEAMing means beads, emulsion, amplification and magnetics. This is a special type of PCR that is highly sensitive and specific. Here, the PCR technology is combined with flow cytometry to identify the specific mutation in DNA along with its quantification.
ARMS is a very helpful technology to detect single base change mutation or deletion of small segment of DNA. In this technique, allele-specific primers are used for DNA amplification.13
Droplet digital PCR (ddPCR) is a novel technology. It precisely quantifies the target nucleic acid with the help of water in oil droplet partition of DNA.
Enhanced ice-cold PCR (E-ice-COLD-PCR) is also a unique technology. It uses a modified blocking oligonucleotide to prevent the amplification of wild-type sequence. The E-ice-COLD-PCR amplifies the mutant sequence of the target DNA13 and, subsequently, the exact identification of the mutant sequence is known as NGS.
4.2 Epigenetic study
DNA hypermethylation in the promoter region can be assessed by methylation-specific PCR protocol. Microarray-based DNA methylation assay technology is very fast and helps in extensive assessment of DNA methylation in cfDNA. NGS in comparison to microarray-based technology covers a wide range of genomic methylation.15 Digital methyl light is a novel technology that can detect a single methylated allele. In addition, droplet digital methyl light is also used, which is more than 25-fold more sensitive than digital methyl light technology.
4.3 Study of RNA
Quantitative reverse transcriptase PCR is the most reliable method for the detection and amplification of microRNA and sRNA. However, microarray and NGS technologies can also be used for the detection and analysis of various types RNA.
4.4 Tumour cell phenotyping and morphology
Tumour cell phenotyping can be studied by using a panel of antibodies consisting of CK, EpCAM, HER-2 etc.
5 CLINICAL APPLICATIONS OF LIQUID BIOPSY
The various clinical applications of liquid biopsy are listed in Table 2.
| Applications | Implementation |
|---|---|
| Early diagnosis and screening of cancer |
|
| Monitoring minimal residual disease |
|
| Management |
|
| Prognostic markers |
|
- CTC, circulating tumour cell.
5.1 Early diagnosis and screening of cancer
- Simple quantitation of cfDNA: Simple quantitative estimation of cfDNA may be helpful to assess the presence of cancer. Several studies have highlighted the presence of a higher amount of cfDNA in the blood of the cancer patients than in that of healthy control. However, many studies have also shown that cfDNA may not be raised in cancer and the simple quantitation of cfDNA may not be a reliable biomarker of cancer.16
- Detection of tumour-specific mutation: Various tumour-specific DNA mutations may be detected in the liquid biopsy sample to indicate the probable presence of cancer. Chen et al17 analysed loss of heterozygosity and microsatellite instability in cfDNA in the plasma of breast carcinoma patients. They detected the alteration of DNA in 48% of the patients including two cases of in situ carcinoma of breast. These are interesting and promising data because they indicate that in situ carcinoma may also show changes in DNA in the liquid biopsy sample. He et al18 noted the mutational changes in cfDNA of non-small cell lung carcinomas and suggested it as an early biomarker of such carcinomas.
- Tumour-specific miRNA and sRNA: miRNA and sRNA can be analysed to detect the early carcinoma cases. Various groups of people analysed miRNA by quantitative real time PCR in plasma samples and suggested that this may be useful to detect an early lung carcinoma patients.19 Roberts et al21 analysed sRNA and miRNA by high-depth small RNA sequencing in plasma of colorectal adenoma patients and claimed that the demonstration of these biomarkers may be important to screen colorectal adenoma patients in future.
5.2 Monitoring minimal residual disease
A liquid biopsy can be used for the detection of minimal residual diseases in various carcinomas including colorectal, lung and breast carcinomas.22 Beaver et al23 studied cfDNA in breast carcinoma cases by ddPCR. They identified PIK3CA exon 20 mutations (H1047R) in both pre- and post-surgical patients and suggested that the detection of these mutations by ddPCR may be helpful to detect minimal residual disease in breast carcinoma.23
6 MANAGEMENT
A liquid biopsy may be useful for the management of different carcinomas. Paoletti et al24 used a panel of antibodies consisting of ER, BCL2, HER2 and Ki67 on the CTCs of breast carcinoma cases and created an endocrine therapy index to manage such patients. It has been noted that the significant number of cases (60%) of non-small cell lung carcinoma show resistance to epidermal growth factor receptor (EGFR) inhibitor therapy due to the development of EGFR T790M mutation. Castellanos-Rizaldos et al25 extracted exosomal RNA/DNA and cfDNA from the plasma of non-small cell lung carcinoma cases and demonstrated T790M mutation by allele-specific qPCR assay. The demonstration of this specific mutation may be helpful to use the third-generation tyrosine kinase inhibitors by simply examining the blood of the patient.
Similarly, mutated KRAS genes in colorectal cancer may be resistant to anti-EGFR therapy and this mutation can be demonstrated by ddPCR. Sclafani et al26 in a recent study detected KRAS/BRAF mutations by ddPCR in circulating tumour DNA (ctDNA) from the blood sample of the colorectal carcinoma patients. Similarly, BRAF mutations have also been demonstrated in CTCs and ctDNA of the blood sample of melanoma patients. This may be helpful for BRAF-targeting therapy in melanoma.27
6.1 Prognostic markers
The analysis of liquid biopsy may be helpful to assess the prognosis of several carcinomas. Rack et al28 demonstrated that the presence of CTCs in early breast carcinoma has poor survival and suggested that the presence of CTCs both before and after adjuvant chemotherapy has a higher risk of relapse in carcinoma of the breast. In fact, increased CTC counts are well correlated with relapse in colorectal cancer, liver cancer, urothelial cell cancer and oesophageal cancer.28, 30, 31
Certain mutations in the tumour, such as KRAS hotspot mutations and cyclin-dependent kinase inhibitor 2A (CDKN2A) hyper methylation, are considered as prognostically bad in patients with colorectal carcinomas. Lecomte et al32 analysed ctDNA for the mutation of KRAS and hyper methylation of CDKN2A in colorectal cancer patients and noted that patients devoid of such mutated DNA in the blood sample had better disease-free survival.
7 DISCUSSION
Liquid biopsy has opened a new horizon in pathology. It is a promising technology and may have a tremendous impact in diagnosis, management and prognosis of the patient. It helps in the diagnosis of the inaccessible tumour particularly in lung carcinomas and colonic carcinomas. ctDNA, CTCs, exosomal miRNA and sRNA provide the valuable information of the primary tumour, and real-time changes of the tumour during therapy (Table 3). Tumour phenotyping and serial mutational changes in the tumour DNA are helpful indicators of targeted tumour therapy. There are several challenges waiting for liquid biopsy. The conventional biopsy is many times cheaper than a liquid biopsy and less complicated. Therefore, the liquid biopsy may not be able to replace the conventional biopsy in the near future. Enrichment and the separation of CTCs cells and cfDNA are the most serious challenges regarding the liquid biopsy. The circulating tumour cells are usually too few and EpCAM on the surface of CTCs may not be expressed due to epithelial-mesenchymal transition. Therefore, at times, it may be more difficult to identify these CTCs in the circulation. Similarly, both normal and tumour cells release DNA in the circulation and so it is necessary to separate the circulating DNA of normal cells from the tumour DNA. Another major challenge of liquid biopsy is the detection of mutations in the very low amount of DNA. The various newer technologies such as ddPCR and BEAMing may be helpful in this aspect. Finally, we should remember that the mutational change may occur in a normal person in an advanced age, and therefore the presence of a mutation may not simply represent the disease. In spite of all these limitations, we still suggest that liquid biopsy is a very promising technology and before the widespread clinical application of liquid biopsy, many more large studies should be done to compare liquid biopsy and conventional biopsy.
| Factors | Liquid biopsy | Conventional biopsy and fine needle aspiration biopsy |
|---|---|---|
| Cost of sample collection | Cheaper to do | More costly |
| Sample preparation | Difficult and needs a sophisticated instrument | Relatively easy |
| Study of tumour heterogeneity | Possible | FNAB may be done from multiple sites and is helpful. However, in the conventional biopsy, it is not possible |
| Real-time monitoring | Possible | Not feasible |
| Minimal residual disease detection | Possible | Not feasible |
| Personalised treatment | Possible | Possible |
| Archival tissue preservation | Not always possible | Possible in conventional biopsy and not possible in FNAB |
- FNAB, fine needle aspiration biopsy.
CONFLICT OF INTEREST
None.