Association of nitric oxide synthase gene polymorphism with asthma: A systematic review and meta-analysis
Abstract
Introduction
This study examines the associations between asthma and nitric oxide (NO) synthase (NOS) gene polymorphisms.
Methods
After a systematic literature search in electronic databases, studies were selected based on eligibility criteria. Data were extracted from research articles and were synthesized and tabulated. Where a particular polymorphism data were reported by multiple studies, meta-analyses of odds ratios were performed, or odds ratios reported by individual studies were pooled.
Results
Twenty studies (4450 asthma patients and 5306 non-asthmatic individuals) were identified. Many studies did not find any association between CCTTT repeat polymorphism in NOS2 gene and asthma. However, a study reported that pretreatment mean exhaled NO levels in asthmatics were found to be significantly higher in genotypes with higher number of CCTTT repeats. Also, alleles with <11 CCTTT repeats were associated with poor asthma treatment outcomes. A single nucleotide polymorphism, G894T, in NOS3 gene was not found to be significantly associated with asthma by at least four studies. However, a T allele at this locus was associated with lower NO levels. Also, G894T frequency was significantly higher in asthmatic children who responded to inhaled corticosteroids along with long-lasting beta2-agonists. A T allele of NOS3 786C/T polymorphism increased the probability of bronchial asthma with comorbid essential hypertension in asthma patients. Asthma severity also differed for different Ser608Leu exon 16 variants of NOS2 gene.
Conclusions
Several polymorph NOS gene variants are identified, some of which appear to have influence on asthma prevalence or outcomes. However, data are varying depending on the nature of variant, ethnicity, study design, and disease parameters.
1 INTRODUCTION
Asthma is a multifactorial chronic inflammatory disease affecting both children and adults. It is the most common chronic disease in children. Asthma is characterized by bronchial constriction and hyperreactivity, airflow limitation, and mucous hypersecretion, which leads to symptoms such as wheezing, coughing, and shortness of breath.1 Increase in secretions, reduced airway pliability, and the presence of mucosal edema may further increase the bronchoconstriction.2 Structural and functional alterations in respiratory epithelium play a crucial role in asthma pathophysiology. Such alterations include airway wall thickening, subepithelial fibrosis, myocyte hypertrophy, myofibroblast hyperplasia, and mucus metaplasia.3
The prevalence of asthma is considerably high. In USA, the prevalence of asthma is 12.8% (https://www.cdc.gov/asthma/nhis/2019/data.htm); whereas in China, its prevalence is estimated at 4.3%, which can be underestimated because of underdiagnosis.4 In 2019, 262 million people suffered from asthma worldwide. Mortality due to asthma is higher in low-income countries where asthma remains underdiagnosed and undertreated (https://www.cdc.gov/asthma/nhis/2019/data.htm; https://www.who.int/news-room/fact-sheets/detail/asthma). Cigarette smoking, allergic rhinitis, childhood pneumonia or bronchitis, parental history of a respiratory disease, and low educational attainment are identified as risk factors for adult asthma.4 Obesity is also an important risk factor for the development of asthma (https://www.cdc.gov/asthma/nhis/2019/data.htm).
Nitric oxide (NO) is a ubiquitous signaling molecule acting as an important vasodilator, neurotransmitter, and inflammatory mediator. However, it also plays a role in the pathophysiology of asthma. NO is synthesized by the NO synthase (NOS) enzymes, which are found to be of three types. NOS1 is mainly expressed by the noradrenergic, non-cholinergic nerve fibers of the airway. NOS2 is mainly found in inflammatory immune cells and is also present in the respiratory epithelium. NOS3 is mainly found in the endothelial cells but it is also observed in the bronchiolar and alveolar cells.5 NO is formed by the NOS enzymes when L-arginine is converted to L-citrulline. NO is a potent molecule for the recruitment of inflammatory cells and for the amplification of inflammatory response.6
One of the major anatomical locations where NO is produced is the paranasal sinuses where it is synthesized by the NOA2A. Whereas NO is constitutively expressed in nasal sinus epithelial surfaces, it is not generally expressed in the nasal cavity.7 Exhaled air of the asthmatic patients contains higher levels of NO that can be reduced upon corticosteroid treatment.6 Exhaled NO acts as an inflammatory biomarker of the airway8 and high levels of exhaled NO can be used to classify asthma severity and helps in identifying patients at risk.9
Asthma predisposition involves gene–environment interactions in its onset as well as severity. Etiology of asthma has genetic elements, and its heritability is reported to be between 36% and 77%. Over 100 genes are implicated for the pathogenesis of asthma and related conditions.10 Polymorphisms are observed in several genes related to asthma pathophysiology, and NOS genes also exhibit multiple variants. The objective of the present study was to conduct a systematic literature search for the identification of the studies that reported the association between a NOS gene variant and asthma prevalence or severity.
2 MATERIALS AND METHODS
2.1 Inclusion and exclusion criteria
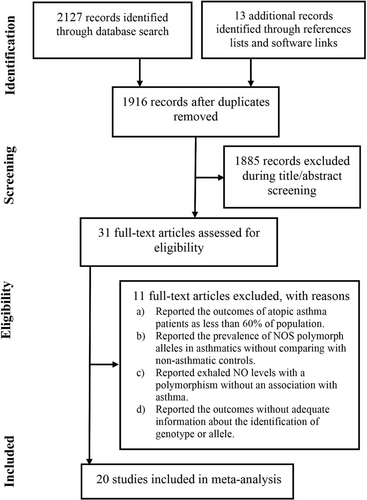
The studies included in this systematic review are those that (a) examined the association between NOS gene polymorphism and asthma incidence or severity; (b) reported statistical indices of the relationship between a gene variant and incidence or severity of asthma; (c) reported incidence of asthma in a cohort of individuals with a particular NOS variant; and (d) reported the association between NOS polymorphism and IgE levels in asthma patients. Studies were, however, excluded if they (a) reported the outcomes of atopic asthma patients but their proportion was less than 60% in the study population; (b) reported the prevalence of NOS polymorph alleles in asthmatics without comparing with non-asthmatic controls; and (c) reported the outcomes without adequate information about the identification of genotype or allele.
2.2 Literature search
The literature search was conducted in electronic databases (Google Scholar, Ovid, PubMed, Science Direct, and Wiley). Most relevant keywords were used for literature search by using these as phrases: asthma, nitric oxide synthase, NOS, NOS1, NOS2, NOS3, inducible, iNOS, endothelial, eNOS, gene polymorphism, variants, genetic variations, repeat, copy, promoter, intron, and phenotype. The literature search encompassed original research articles published from the data of database inception until October 2021. After the identification of relevant research articles, the references lists of these articles were also screened for additional records.
2.3 Data synthesis and analyses
Demographic, allelic or genotypic, polymorph or variant data, prevalence in asthmatics and non-asthmatics, associational statistics including odds ratios and regression coefficients, and immunoglobin E (IgE) levels in asthmatics with a particular allele variant were extracted from the research articles of respective studies and synthesized according to the type of NOS or the presence of polymorphism. For studies that reported the descriptive data regarding the prevalence of polymorphism in association with asthma or its severity, odds ratios were calculated. For polymorphic genes whose outcomes were reported by more than one study, the odds ratios were pooled using DerSimonian–Liard method to achieve overall estimates.
3 RESULTS
Twenty studies were identified from the literature by following the eligibility criteria.11-30 A flowchart of study screening and selection process is presented in Figure 1. In these studies, 4450 asthmatic patients and 5306 non-asthmatics were recruited and studied for NOS polymorphism. The age of asthma patients were 29.9 years [95% confidence interval (CI): 23.2, 36.5] (range 8.5 ± 0.2 to 60 ± 1.1) and 47% [95% CI: 40, 53] of these patients were females.
Several authors have reported the odds ratios depicting an association between a NOS gene polymorphism and asthma risk. Table 1 shows NOS gene polymorphisms, many of which had a statistically significant association with asthma. Several studies did not report the odds ratios of the association between a polymorphic gene and asthma risk but provided numeric data showing the prevalence of asthma in carriers and noncarriers. For these studies, odds ratios were calculated from raw data and are presented in Table 2.
| Study | Polymorph allele | Association | OR [95% CI]; p-value |
|---|---|---|---|
| NOS1 | |||
| Gao 2000 | Homozygous 183-bp alleles with VNTR in intron 2 | Higher risk of asthma vs controls in carriers | 2.08 [1.2, 3.57]; p = 0.01 |
| Grasemann 2000 | Allele with 17 CA-repeat in exon 29 | Higher risk of asthma vs controls in carriers | 1.49 [1.17, 1.9]; p = 0.0013 |
| Grasemann 2000 | Allele with 18 CA repeat in exon 29 | Lower risk of asthma vs controls in carriers | 0.49 [0.3, 0.8]; p = 0.0006 |
| Grasemann 2000 | Allele with 17 CA repeat in exon 29 | Risk of high IgE levels in asthmatic carriers | 8.2 [5.93, 11.3]; p < 0.0001 |
| Grasemann 2000 | Allele with 18 CA repeat in exon 29 | Risk of high IgE levels in asthmatic carriers | 8.15 [5.9, 11.3]; p < 0.0001 |
| Holla 2004 | SNP with C/T transition located 276-bp downstream of translation termination site (C5266T) in exon 29 | Higher risk of serum IgE levels in T allele carriers | 2.08 [1.52, 2.85]; p < 0.01 |
| Wechsler 2000 | Alleles with >12 AAT repeats | Lower risk of asthma in carriers | 0.62 [0.43, 0.89]; p = 0.01 |
| NOS2 | |||
| Gao 2000 | Homozygous Glu298Asp | No significant risk of asthma | 0.73 [0.46, 1.17]; p = 0.19 |
| Batra 2007 | Allele with 3 GT repeats in intron 4 | Higher risk of severe asthma in carriers | 2.62 [1.03, 6.6]; p = 0.04 |
| Hirai 2018 | Alleles with <11 CCTTT repeats | Higher risk of asthma exacerbations in carriers | 2.8 [1.2, 6.6]; p = 0.016 |
| Holla 2006 | T allele with Ser608Leu polymorphism in exon 1 | Higher risk of asthma severity in carriers | 5 [1.88, 13.3]; p = 0.0005 |
| NOS3 | |||
| Gao 2000 | Homozygous 311 bp allele with 5′ promoter VNTR | No significant risk of asthma | 1.07 [0.6, 1.92]; p = 0.81 |
| Bouzigon 2012 | rs743507 (Major allele T and minor allele C) | Higher risk of high FeNO in asthmatics | Beta coefficient 0.08 [0.03, 0.14]; p = 0.004 |
| Holla 2008 | -786rs2070744 | Higher risk of asthma in CC vs TT genotype | 1.6 [0.97, 2.66]; p = 0.028 |
| Holla 2008 | -786rs2070744 | Higher risk of asthma CT vs TT genotype | 1.27 [0.9, 1.8]; p = 0.028 |
| Holla 2008 | -691rs3918226 | Higher risk of asthma CT vs CC genotype | 1.67 1.05, 2.64]; p = 0.039 |
| Holla 2008 | G11Trs1799985 | Lower risk of asthma GT vs GG genotype | 0.81 [0.57, 1.14]; p = 0.039 |
| Lower risk of asthma TT vs GG genotype | 0.67 [0.42, 1.1]; p = 0.043 | ||
| Holla 2008 | 27-bp repeat | No significant risk of asthma aa vs bb genotypes | 1.2 [0.52, 2.78] |
| No significant risk of asthma ab vs bb genotypes | 1.05 [0.74, 1.49] | ||
| Holla 2008 | 774rs1549758 | No significant risk of asthma CT vs CC genotypes | 1.13 [0.81, 1.57] |
| No significant risk of asthma TT vs CC genotypes | 1.09 [0.57. 2.06] | ||
| Holla 2008 | 894rs1799983 | No significant risk of asthma GT vs GG genotypes | 1.18 [0.85, 1.65] |
| No significant risk of asthma TT vs GG genotypes | 1.22 [0.68, 2.19] | ||
| Shakhanov 2017 | 786C/T | Higher risk of comorbid asthma and essential hypertension compared with bronchial asthma alone | 2.40 [1.04, 5.56] |
| van's Gravesande 2003 | G894T | No significant risk of asthma | 1.07 [0.51, 2.25]; p = 0.86 |
| van's Gravesande 2003 | G894T | No significant risk of asthma | 2.50 [0.61, 10.3]; p = 0.2 |
- Abbreviations: IgE, immunoglobin E; NOS, nitric oxide synthase, OR, odds ratio.
| Study | Polymorphic allele | Genotype | Asthmatics | Non-asthmatics | Odds ratio [95% CI]; p-value | ||
|---|---|---|---|---|---|---|---|
| Carriers | Noncarriers | Carriers | Noncarriers | ||||
| NOS1 | |||||||
| Gao 2000 | 183-bp allele with VNTR in intron 2 | Homozygous 183-bp | 62 | 24 | 119 | 95 | 2.06 [1.2, 3.55]; p = 0.009 |
| Holla 2004 | C/T polymorph in exon 29 | CC | 83 | 174 | 67 | 146 | 1.04 [0.7, 1.54]; p = 0.846 |
| CT | 56 | 120 | 94 | 200 | 0.99 [0.66, 1.48]; p = 0.972 | ||
| TT | 11 | 26 | 139 | 294 | 0.89 [0.43, 1.86]; p = 0.767 | ||
| Leung 2005 | AAT-repeat | >12/>12 repeats | 49 | 42 | 52 | 58 | 1.3 [0.75, 2.27]; p = 0.354 |
| Heterozygous | 15 | 20 | 86 | 80 | 0.7 [0.33, 1.46]; p = 0.337 | ||
| <12/<12 repeats | 37 | 38 | 64 | 62 | 0.94 [0.53, 1.67]; p = 0.841 | ||
| Leung 2005 | C5266T SNP | CC | 45 | 42 | 56 | 58 | 1.11 [0.63, 1.94]; p = 0.715 |
| CT | 44 | 44 | 57 | 56 | 0.98 [0.56, 1.72]; p = 0.95 | ||
| TT | 11 | 14 | 90 | 86 | 0.75 [0.32, 1.74]; p = 0.505 | ||
| NOS2 | |||||||
| Gao 2000 | 311 bp allele with 5′ promoter VNTR | Homozygous 311-bp | 144 | 96 | 37 | 23 | 0.93 [0.52, 1.67]; p = 0.813 |
| Konno 2001 | CCTTT-repeat | Non-14/non-14 repeat | 229 | 25 | 220 | 23 | 0.96 [0.53, 1.74]; p = 0.887 |
| Heterozygous | 22 | 232 | 22 | 221 | 0.95 [0.51, 1.77]; p = 0.878 | ||
| 14/14 repeat | 3 | 251 | 1 | 242 | 2.89 [0.30, 28.0]; p = 0.359 | ||
| Leung 2006 | CCTTT-repeat | Non-14/non-14 repeat | 241 | 143 | 50 | 29 | 0.98 [0.59, 1.61]; p = 0.929 |
| Heterozygous | 49 | 27 | 242 | 145 | 1.09 [0.65, 1.82]; p = 0.749 | ||
| 14/14 repeat | 1 | 2 | 290 | 170 | 0.29 [0.03, 3.26]; p = 0.318 | ||
| NOS3 | |||||||
| Gao 2000 | Glu298Asp | Homozygous Glu | 96 | 54 | 85 | 65 | 1.36 [0.85, 2.16]; p = 0.195 |
| Holla 2002 | A549G | AA | 87 | 111 | 39 | 41 | 0.82 [0.49, 1.39]; p = 0.466 |
| AG | 35 | 39 | 91 | 113 | 1.11 [0.65, 1.90]; p = 0.691 | ||
| GG | 4 | 2 | 122 | 150 | 2.46 [0.44, 13.7]; p = 0.304 | ||
| Holla 2008 | -691rs3918226 | CC | 241 | 278 | 53 | 38 | 0.62 [0.4, 0.98]; p = 0.039 |
| CT | 52 | 36 | 242 | 280 | 1.67 [1.06, 2.64]; p = 0.028 | ||
| TT | 1 | 2 | 293 | 314 | 0.54 [0.05, 5.94]; p = 0.611 | ||
| Holla 2008 | 774rs1549758 | CC | 145 | 165 | 149 | 151 | 0.89 [0.65, 1.22]; p = 0.475 |
| CT | 128 | 129 | 166 | 187 | 1.12 [0.81, 1.54]; p = 0.498 | ||
| TT | 21 | 22 | 273 | 294 | 1.03 [0.55, 1.91]; p = 0.931 | ||
| Holla 2008 | G11Trs1799985 | GG | 131 | 121 | 163 | 195 | 1.30 [0.94, 1.79]; p = 0.117 |
| GT | 125 | 143 | 169 | 173 | 0.89 [0.65, 1.23]; p = 0.496 | ||
| TT | 38 | 52 | 256 | 264 | 0.75 [0.48, 1.18]; p = 0.22 | ||
| Lee 2000 | Endothelial constitutive NOS | bb | 91 | 272 | 30 | 38 | 0.42 [0.25, 0.72]; p = 0.002 |
| ab | 29 | 35 | 92 | 275 | 2.48 [1.43, 4.27]; p = 0.001 | ||
| aa | 1 | 3 | 120 | 307 | 0.85 [0.09, 8.28]; p = 0.891 | ||
| Yanamandra 2005 | VNTR in intron 4 | aa | 7 | 18 | 107 | 318 | 1.16 [0.47, 2.84]; p = 0.753 |
| bb | 60 | 196 | 54 | 140 | 0.79 [0.52, 1.22]; p = 0.289 | ||
| cc | 0 | 0 | 114 | 336 | |||
| ab | 36 | 103 | 78 | 233 | 1.04 [0.66, 1.65]; p = 0.854 | ||
| ac | 7 | 5 | 107 | 331 | 4.33 [1.35, 13.9]; p = 0.014 | ||
| bc | 6 | 14 | 108 | 322 | 1.28 [0.48, 3.41]; p = 0.624 | ||
- Abbreviation: NOS, nitric oxide synthase.
A very few gene polymorphisms were reported by more than one individual study. A meta-analysis of four studies found that G894T variant of NOS3 gene (Glu298Asp) was not found to be significantly associated with asthma risk (Figure 2). A meta-analysis of two studies found that a polymorphism of NOS3, −786 T/C, was also not significantly associated with asthma risk (Figure 2). Another meta-analysis of two studies found that a 27-bp repeat polymorphism of NOS3 was also not significantly associated with asthma risk (Figure 2).
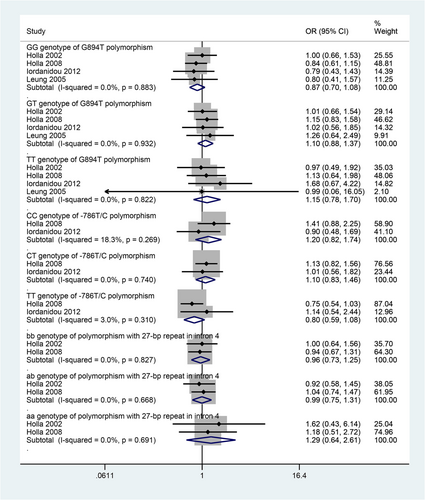
Table 3 presents the outcomes of the studies that reported the association between NOS gene polymorphism and IgE levels in asthma patients. In asthmatics, a polymorphism in exon 29 of NOS1 gene (homozygous for C allele) was associated with lower IgE levels. A homozygous genotype for <12 AAT repeat was found to be associated with significantly lower IgE levels; whereas, a heterozygous genotype (<12 and >12 AAT repeat) was significantly associated with higher IgE levels in asthmatic patients.
| Study | Polymorphic allele | Genotype | High IgE levels | Low IgE levels | Odds ratio [95% CI]; p-value | ||
|---|---|---|---|---|---|---|---|
| Carriers | Noncarriers | Carriers | Noncarriers | ||||
| NOS1 | |||||||
| Gao 2000 | 183-bp allele with VNTR in intron 2 | Homozygous 183-bp | 45 | 88 | 41 | 126 | 1.572 [0.95, 2.60]; p = 0.078 |
| Holla 2004 | C/T polymorphism in exon 29 | CC | 71 | 78 | 129 | 87 | 0.614 [0.40, 0.94]; p = 0.023 |
| CT | 61 | 88 | 73 | 143 | 1.358 [0.88, 2.09]; p = 0.165 | ||
| TT | 17 | 132 | 14 | 202 | 1.858 [0.89, 3.90]; p = 0.101 | ||
| Leung 2005 | C5266T | CC | 124 | 171 | 80 | 94 | 0.852 [0.58, 1.24]; p = 0.406 |
| CT | 127 | 168 | 78 | 96 | 0.930 [0.64, 1.36]; p = 0.708 | ||
| TT | 41 | 254 | 16 | 158 | 1.594 [0.87, 2.94]; p = 0.135 | ||
| Leung 2005 | AAT-repeats | <12/<12 repeats | 41 | 254 | 38 | 136 | 0.578 [0.36, 0.94]; p = 0.028 |
| Heterozygous | 148 | 147 | 68 | 106 | 1.569 [1.07, 2.30]; p = 0.02 | ||
| >12/>12 repeats | 109 | 186 | 70 | 104 | 0.871 [0.59, 1.28]; p = 0.48 | ||
| NOS2 | |||||||
| Gao 2000 | 311 bp allele with 5′ promoter VNTR | Homozygous 311-bp | 105 | 28 | 135 | 32 | 0.889 [0.50, 1.57]; p = 0.684 |
| Leung 2006 | CCTTT-repeat | non-14/non-14 repeat | 231 | 60 | 148 | 24 | 0.624 [0.37, 1.05]; p = 0.074 |
| Heterozygous | 45 | 246 | 31 | 141 | 0.832 [0.50, 1.38]; p = 0.473 | ||
| 14/14 repeat | 1 | 290 | 2 | 170 | 0.293 [0.03, 3.26]; p = 0.318 | ||
| NOS3 | |||||||
| Gao 2000 | Glu298Asp | Homozygous Glu | 67 | 66 | 83 | 84 | 1.027 [0.65, 1.62]; p = 0.907 |
| Leung 2005 | G894T | GG | 227 | 68 | 138 | 36 | 0.871 [0.55, 1.37]; p = 0.552 |
| GT | 65 | 230 | 35 | 139 | 1.122 [0.71, 1.78]; p = 0.624 | ||
| TT | 3 | 292 | 2 | 172 | 0.884 [0.15, 5.34]; p = 0.893 | ||
- Abbreviations: IgE, immunoglobin E; NOS, nitric oxide synthase, VNTR, variable number of tandem repeats.
4 DISCUSSION
In this systematic review, we have identified 21 studies that evaluated the association between asthma and one or more polymorphisms in a NOS gene. Several NOS gene polymorphisms are found to have significant association with asthma. Moreover, a few polymorphisms were found to have significant associations with IgE levels in asthma patients. However, many of the polymorphisms were reported only by a single study.
A number of studies have evaluated the role of a polymorphism in NOS2 gene characterized by the CCTTT repeats. Konno et al (2001) described 14-repeat CCTTT allele as a potentially susceptible or disease modifying allele of inflammatory immune diseases such as atopy. They found significantly higher prevalence of the 14-repeat CCTTT allele in the NOS2 promoter in non-atopic individuals (odds ratio [OR] for the presence of atopy between carriers and non-carriers was 0.42 [95%CI: 0.23, 0.79]). The OR for the development of atopy was independent of asthma, as the genetic effect of the 14-repeat CCTTT allele was persistent even after controlling the age, sex, smoking, and asthma status. Atopic asthma patients constituted 66% of this population.22 Leung et al (2006) also found no association between CCTTT repeats and asthma or exhaled NO in Chinese asthma patients.25
Batra et al (2007) did not find any significant association between 14-repeat CCTTT allele with asthma in Indian asthma patients.11 Pascual et al (2008) also did not find any association between CCTTT repeats in the NOS2A and atopic asthma in a Spanish population, in which increasing number of CCTTT repeat was associated with nasal polyposis instead.26 However, Sato et al (2016) found that the mean exhaled NO levels before treatment in Japanese asthmatic patients were significantly higher in genotypes with higher number of CCTTT repeats (ranged between 9 and 20) in NOS2A proximal promoter pentanucleotide microsatellite.31 In Japanese asthma patients, Hirai et al (2018) suggested that alleles with <11 repeats of CCTTT contribute to poor asthma treatment outcomes when they found that carriers of alleles with <11 repeats were at a higher risk of asthma exacerbation. However, a 12-repeat CCTTT polymorphism was found to be associated with high serum total IgE levels and serum NO levels.15
A single nucleotide polymorphism, the G894T, in the NOS3 gene has been described in asthmatic patients by at least five authors. In the present study, a meta-analysis of four studies also could not find a significant association between G896T polymorphism and asthma. Holla et al (2002) studied atopic asthma in Czech population, and Gao et al (2000) studied in British population.13, 16 Both these authors found no significant association between NOS G894T polymorphism and asthma. van's Gravesande et al (2003) also did not find a significant association between G894T mutation and asthma. There was also no association between G894T mutation and either FEV1 or change in FEV1 post-albuterol treatment in this study. However, T allele at this locus was associated with lower NO levels.28 Iordanidou et al (2012), who also did not find a significant association between G894T polymorphism and asthma, found that the G894T frequency was significantly higher in asthmatic children who responded to inhaled corticosteroids along with long-lasting beta2-agonists in comparison with non-responders.20
Among the other polymorphisms, there was no significant association between asthma and a single nucleotide polymorphism in NOS3 gene, the −786 T/C, as observed by two studies in Czech and Greek Caucasian patients.19, 20 The −786 T/C polymorphism in NOS3 retards the transcription of NOS3 gene, which causes reduction in NO production up to 40%.32 Shakhanov et al (2007) reported that T allele of NOS3 −786C/T polymorphism increased the probability of bronchial asthma with comorbid essential hypertension 2.4 times in Russian asthma patients.27
Grasemann et al (2000) found a significantly decreased risk of asthma with NOS1 allele-18 containing polymorphism in exon 29 but an increased risk with allele 17 at this locus in the Caucasian patients residing in the USA. They found no association between these mutations and IgE levels.14 Gao et al (2000) reported that in NOS1 microsatellite, homozygous 183-bp alleles were significantly associated with asthma but not with atopy in British patients.13 Wechsler et al (2000), who studied a cohort of asthmatic patients consisting mainly of US white individuals, speculated that asthmatics harboring two alleles with 12 or more AAT repeats in NOS1 gene may have diminished exhaled NO levels.29 NO levels are found variable depending on the presence of single nucleotide polymorphisms in NOS genes.33
Yanamandra et al suggested that the variable tandem number of repeats in intron 4 of NOS3 appears to be a risk factor for asthma development in Caucasians or African Americans residing in the USA.30 Holla et al (2006) found that the asthma severity differed for different Ser608Leu exon 16 variants of NOS2 gene in Czech patients. Forty-one percent patients with mild to moderate asthma had more active T allele of Ser608Leu; whereas, only 12% patients with intermittent form of asthma had this allele.18 Batra et al (2007) found a significant association of allele 3 with 15-repeat GT in intron 4 of NOS2A gene with higher risk of asthma, asthma severity, and eosinophil percentage in peripheral blood in Indian asthma patients.11
Taken together, several lines of evidence suggest the role of polymorphisms in NOS genes to affect asthma etiology or prognosis. However, data are varying depending on the nature of variant, ethnicity, study and comparison design differences, and disease parameters. Moreover, a very few polymorphisms are studied in different ethnic groups or localities. Thus, replication of available data will refine the evidence regarding the role of various NOS gene polymorphisms in affecting the etiology and prognosis of asthma.
AUTHOR CONTRIBUTIONS
Zeru Fan wrote the manuscript; Tao Liu and Wei Na collected and analyzed the data. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST STATEMENT
None.
ETHICS STATEMENT
N/A
Open Research
DATA AVAILABILITY STATEMENT
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.




