Role of epigenetic modifications and aging in inflammatory bowel disease
Abstract
Inflammatory bowel disease (IBD), which encompasses ulcerative colitis (UC) and Crohn's disease (CD), refers to a chronic and recurrent nonspecific inflammatory condition affecting the mucosal and submucosal layers of the intestines. A positive family history has been identified as a risk factor for the onset of IBD, likely influenced by genetic and environmental factors. In addition to intestinal damage, patients may have extraintestinal manifestations such as inflammation of the skin, eyes, and joints, inflammation of the liver or bile ducts, kidney stones, iron-deficiency anemia, and growth retardation in children, which may complicate diagnosis and treatment. Therefore, investigating the mechanisms of IBD and finding precise therapeutic targets provides enormous benefits to patients with IBD. Multiple studies have consistently demonstrated the influential role of dietary metabolism and aging in the development of IBD. Moreover, emerging evidence suggests that aging often coincides with alterations in epigenetic modifications, while diet metabolism mediates these epigenetic changes. Epigenetics has emerged as a prospective field for identifying novel biomarkers to facilitate the diagnosis, prognosis, and treatment of diseases. Therefore, in this perspective, we summarize the cross-talk between epigenetic modifications, diet metabolism, and aging in the pathogenesis of IBD and attempt to identify new potential therapeutic targets and strategies.
1 INTRODUCTION
Inflammatory bowel disease (IBD), which comprises Crohn's disease (CD) and ulcerative colitis (UC), refers to chronic inflammatory conditions affecting the gastrointestinal tract. CD affects the entire gut, while UC is limited to a specific portion of the gut.1 Furthermore, as IBD advances, there is a potential for the development of colorectal cancer, bowel obstruction, and bleeding.2 The global prevalence of IBD is increasing due to population aging and shifts in dietary patterns. Currently, the diagnosis of IBD and differentiation from other types of inflammation relies on colonoscopy and biopsy. However, surgery is the primary treatment modality, given the insufficiently understood mechanisms underlying IBD pathogenesis.3 Consequently, conducting in-depth research on the mechanisms of IBD is imperative to discover effective prevention and treatment strategies.
The prevalence of IBD has been rapidly increasing and is believed to result from the interplay between genetic factors and environmental influences. These interactions significantly affect the epigenetic modifications that occur in the genome. Epigenetic modifications, reversible and heritable changes to the function of genes, play a crucial role in the development of IBD. Such modifications include DNA methylation, N6-methyladenosine (m6A) of RNA, and posttranslational methylation of histones.4-6 Collectively, these epigenetic modifications target key signaling pathways involved in the pathogenesis of IBD.
Aging is a natural phenomenon characterized by a gradual decline in physiological and psychological adaptations to the environment, ultimately leading to mortality. Notably, there has been a notable rise in the incidence of IBD among individuals aged 60 and above, accounting for 25% of all cases.7 Extensive research has indicated that this increased prevalence in older adults can be attributed to epigenetic modifications, some of which may disrupt dietary metabolism, particularly in a Western diet.8, 9 Moreover, the aging process affects the immune system, resulting in progressive immune system disorders that can lead to mild or even moderate chronic inflammation of the intestines. However, our understanding of the therapeutic aspects of IBD, particularly in older patients, remains limited.
From this perspective, we examined epigenetic modifications and their role in combining two significant risk factors for IBD: dietary metabolism and aging. We aimed to identify the crucial factor in IBD's pathogenesis and propose potential therapeutic targets and strategies.
2 EPIGENETIC MODIFICATIONS IN IBD
Epigenetic modification pertains to regulating gene expression without altering the genetic information. This process, which is genetic, reversible, and dynamic, is particularly conducive to participating in cell development, differentiation, and functionality. Consequently, it can serve as a significant and promising therapeutic target.10, 11 The most prevalent epigenetic alterations include the methylation modifications of DNA, RNA, and histones.12 The catalytic process involved in methylation modification primarily centers on transferring methyl groups from active methyl compounds to other inert compounds. Several methylated sites found in CD are implicated in immune system regulatory pathways, including the macrophage stimulating protein 1 receptor, interleukin-16 (IL-16), IL-10, and leukemia inhibitory factor.13 The process of methylated modification is dynamic and reversible, primarily influenced by the activities of “writers,” “erasers,” and “readers.” These writers function as adenosine methyltransferases, erasers act as demethylases, and readers serve as m6A binding proteins.14 Numerous studies have demonstrated the extensive utilization of epigenetic modifications in treating IBD.
2.1 DNA epigenetic modifications in IBD
DNA methylation is commonly recognized as the most stable and easily accessible form of epigenetic modification. Epigenome-wide association studies have successfully established meaningful links between DNA methylation and the regulation of gene expression in individuals with IBD.15, 16 Multiple studies have provided evidence indicating the involvement of candidate methylated genes in the regulation of innate immune function in patients with IBD. Notably, genes such as interferon-gamma (IFNG), recombinant protease-activated receptor 2 (PAR2), Runt-related transcription factor 3 (RUNX3), estrogen receptor (ER), and interferon regulatory factor 5 (IRF5) have been identified as potential candidates.17-19 However, it is essential to note that only a limited number of these candidate genes have been definitively associated with IBD.
Clinical studies have demonstrated a downward trend in the DNA methylation modifications aimed at safeguarding the regular expression of IFNG, which is disrupted in patients with IBD.18 The suggested DNA methylation patterns of IFNG have a potential role in regulating the pathogenesis of IBD. Multiple studies have consistently reported an association between elevated levels of DNA methylation in the PAR2 gene and the manifestation of a severe phenotype of UC.17 Furthermore, it has been observed that activation of PAR2 in the colon exerts a proinflammatory influence, leading to the generation of T-helper cell type 1 (Th1) cytokines such as tumor necrosis factor-α (TNF-α), IL-1, interferon-γ (IFN-G), and the release of calcitonin gene-related peptide.20 In the context of IBD, it is worth noting that TNF-α, a pivotal mediator, upregulates PAR2, leading to the activation of cytosolic phospholipase A2 as well as the expansion of intestinal myofibroblasts, thus worsening the severity of IBD21 (Figure 1).
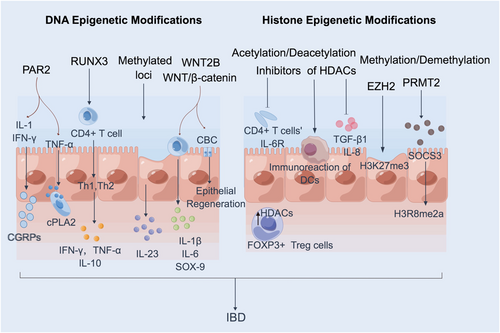
Additionally, RUNX3, a Runt structural domain transcription factor family member, has emerged as a significant regulator of lineage-specific gene expression and has recently been implicated in human autoimmune disorders. The transcription factor RUNX3 has been found to negatively impact the immune response, leading to an increased Th1 or Th2 response. This, in turn, stimulates the expression of immune factors such as TNF-α, interferon-γ (IFN-γ), and IL-10, ultimately exacerbating the severity of IBD22 (Figure 1). The levels of methylation modification in the ER gene were higher in patients with IBD, suggesting that methylation may serve as a potential diagnostic marker for IBD.23 In contrast, analysis of IBD patients found no evidence of DNA methylation in the IRF5 promoter region, indicating that IRF5 is unlikely to play a role in the development of IBD.19 Additionally, the dysregulation of IFN-γ, caused by increased DNA methylation of the IFN-γ gene in T cells from peripheral blood of IBD patients, contributed to IBD's exacerbation.24
In addition, alterations in DNA methylation at specific CpG sites have been observed in individuals with IBD. Notably, changes were detected in several genes involved in the IL-23 pathway, including B-cell lymphoma 3 protein (BCL3), signal transducer and activator of transcription 3 (STAT3), oncostatin-M (OSM), and signal transducer and activator of transcription 5 (STAT5)25 (Figure 1). Moreover, the secretion of recombinant wingless type MMTV integration site family, member 2B (WNT2B) and other canonical WNT ligands has been shown to activate WNT/β-catenin signaling during intestinal inflammation, leading to the release of inflammatory factors from crypt basal columnar stem cells and promoting epithelial regeneration after tissue injury26 (Figure 1). Hence, specific DNA methylation patterns may serve as biomarkers for disease activity in IBD patients and represent novel targets for therapeutic intervention.
2.2 Histone epigenetic modifications in IBD
Histones play a crucial role in chromatin structure and gene expression, with the significant types being H1, H2A, H2B, H3, and H4. These histones, which are integral components of chromatin, participate in regulating gene expression. Posttranslational modifications of histones, specifically those occurring on lysine residues in the tails of unstructured histones protruding from nucleosomes, are commonly known as “epigenetic marks.” These marks confer gene-expression characteristics independent of the DNA sequence.27 In intestinal immune homeostasis, epigenetic histone modification factors play a significant role, with acetylation and methylation being the most prevalent posttranslational epigenetic modifications.
2.2.1 Histone acetylation modifications in IBD
Histone acetylation, an essential histone modification in cellular processes, is regulated by the enzymes histone acetyltransferase (HAT) and histone deacetylase (HDAC). HAT and HDAC function to add or remove acetyl groups to various residues in the histone tails.28 These modifications play a crucial role in the epigenetic regulation of the immune system, influencing the differentiation and function of essential cell types such as monocytes/macrophages, dendritic cells (DCs), neutrophils, and Th cell subpopulations.
There has been a resurgence of interest in butyrate, a type of short-chain fatty acid and HDAC inhibitor, which is produced by the gut microbiota and plays a crucial role in maintaining gut homeostasis.29 Moreover, numerous studies have demonstrated that HDAC inhibitors possess anti-inflammatory properties in intestinal diseases by targeting T cells and macrophages. For instance, in an experimental colitis model, the novel HDAC inhibitor ITF2357 effectively impeded tumor growth in size and number, reduced inflammatory symptoms by decreasing IFN-γ secretion, and increased IL-10 production.30 By treating naïve CD4+ T cells with HDAC inhibitors, chromatin acetylation of the IL-6 receptor (IL-6R) gene and its promoter can be modified. This modification decreases messenger RNA (mRNA) and protein levels of IL-6R in patients with IBD31, 32 (Figure 1).
Moreover, reducing IL-6R activity decreases STAT3 phosphorylation, a downstream effect of IL-6R signaling. Therefore, the IL-6/STAT3/IL-17 pathway is believed to play a crucial role in the anti-inflammatory effect of HDAC inhibitors in experimental colitis models.33 These findings highlight the potential of HDAC inhibition as a therapeutic approach for intestinal diseases. Notably, IBD is characterized by an upregulation of HDAC expression locally, while Foxp3+ T-regulatory cells (Tregs) use HDACs to regulate chromatin remodeling and gene expression31 (Figure 1). Thus, treating IBD may involve targeting Treg functions.
The study found that most inflammation-induced HDACs were significantly downregulated in patients with active IBD compared to those with UC and CD in remission. This indicates the existence of an antagonistic mechanism for localized epithelial tissue damage. Furthermore, the study proposes that pan-HDAC inhibitors, such as givinostat and vorinostat, could potentially suppress intestinal inflammation by inhibiting the secretion of autocrine epithelial transforming growth factor-β1 and reducing the expression of IL-834, 35 (Figure 1). Additionally, the study suggests that HDAC inhibitor-induced epigenetic modifications can shift the function of DC from immunostimulation to immunomodulation, thus providing relief for IBD to some extent36 (Figure 1). In conclusion, the findings support that inhibiting histone deacetylation can reduce IBD by promoting immune tolerance.
2.2.2 Histone methylation modifications in IBD
Histone methylation is crucial in regulating transcriptional activation or repression, primarily determined by the methylation levels and specific residues involved. Enhancer of zest homolog 2 (EZH2), a member of the SET1 methyltransferase family, acts as the catalytic component of polycomb repressive complex 2. Specifically, when EZH2 is overexpressed, it increases histone 3 lysine 27 trimethylations (H3K27me3), a repressive marker. This elevated H3K27me3 level results in silencing inhibitory genes in aberrant cells. Notably, inhibition of EZH2 activity has been shown to alleviate IBD and delay the onset of colitis-associated cancer37 (Figure 1). Protein arginine methyltransferase 2 (PRMT2) plays a critical role in various biological processes, such as gene transcription, mRNA splicing, cell proliferation, and cell differentiation, through its involvement in histone methylation. It has been demonstrated that PRMT2 contributes to IBD by promoting the increased presence of the inhibitory histone mark H3R8 asymmetric methylation in the promoter region of suppressor of cytokine signal transduction 3. Knocking down PRMT2 alleviates dextran sodium sulfate-induced IBD38 (Figure 1). Therefore, interfering with the activity of histone regulators presents a promising avenue for developing novel therapeutic strategies against IBD.
2.3 RNA epigenetic modifications in IBD
Although DNA methylation was initially identified, RNA methylation modifications have recently become a research hotspot. The most common modification is m6A, which regulates RNA function and metabolism.
In recent years, studies of m6A have focused on the m6A writer complex, which consists of methyltransferase-like 3 (METTL3), methyltransferase-like 14 (METTL14), and Wilms tumor 1-associated protein (WTAP).39 Mainly, METTL14 serves as a mediating platform that binds RNA in positively charged grooves formed at its protein interface, thereby increasing the affinity of methyl groups for substrate RNA molecules or promoting RNA binding to substrates. METTL3 accommodates S-adenosyl-l-methionine (SAM) cofactors and transfers methyl groups from SAM to the adenine portion of the receptor. The primary function of WTAP is the recruitment of METTL3 and METTL14.39
Regarding independent m6A modification factors, the mediation by METTL3 promotes immune function and activates the nuclear factor-κB (NF-κB) pathway. In this context, it is referred to as the M6A-XPO1-NF-κB signaling pathway. This pathway involves an increased number of exportin 1 (XPO1) proteins, which stimulate the nuclear factor NF-κB in response to the high levels of m6A methylation at the 5′-untranslated region of XPO1 RNA. Consequently, this activation triggers an inflammatory response that further exacerbates IBD40 (Figure 2). Goh et al. observed that mice deficient in METTL14, predominantly distributed in immune cells, developed spontaneous colitis.41 METTL14 activates the NF-κB pathway by recognizing Toll-like receptors on the surface of DCs, inducing the expression of inflammatory factors such as IL-6, IL-1β, and TNF-α. This activation leads to the dysfunction of intestinal Treg cells, disrupting intestinal immune homeostasis and ultimately contributing to the development of IBD41 (Figure 2). The YT521-B homology domain family protein 1 (YTHDF1) was downregulated in UC patients compared to controls. Mechanistically, YTHDF1 amplifies Wnt/β-catenin signaling at the translational level while maintaining intestinal stem cell regeneration.42 Specifically, YTHDF1 can mitigate IBD by enhancing the translation of lysosomal histone cathepsin in DC and promoting the translation of Wnt signaling effectors, such as transcription factor 7-like 2/T-cell-specific transcription factor 4 (TCF7L2/TCF4).42 Deletion of YTHDF1 in DC induces CD8+ T cells to stimulate high expression of programmed cell death 1 ligand 1 (PD-L1) and IFN-γ, ultimately exacerbating IBD42 (Figure 2). It was observed that in tumor necrosis factor (TNF)-stimulated fibroblasts, inhibition of the expression of histone lysine-specific demethylase 4A (KDM4A), a demethylase of histone 3 lysine 9 trimethylation (H3K9me3) on the IL-6 promoter, or induction of trimethylation of histone 3 lysine 9 (H3K9), weakened the binding between NF-κB and the IL-6 promoter, subsequently alleviating IBD.43 Moreover, the intestinal inflammatory response disrupts the intestinal mucosal barrier and the intestinal microbiota, thereby increasing the likelihood of exacerbating IBD44 (Figure 2). Nevertheless, owing to the instability of m6A modification, its suitability as the gold standard for clinical diagnosis and targeted therapy of IBD remains controversial.

3 AGING IN IBD
As an intricate natural phenomenon, aging represents a spontaneous and inevitable process within an individual's life. Aging and IBD are intricately intertwined, with approximately one in 160 older adults afflicted by IBD. The prevalence of IBD in older adults appears to be escalating at 5.2% per year. Alarmingly, up to 15% of IBD patients in North America and Asia are diagnosed after reaching 60. The incidence of IBD among individuals aged over 60 is estimated to be four to eight cases per 100,000 individuals.45 Considering the prevalence of IBD in the elderly and the aging trajectory of IBD patients, there is an anticipated surge in the number of elderly individuals affected by IBD. The care of elderly patients with IBD poses distinctive challenges regarding diagnosis and management decision-making.
3.1 Age-related diseases in IBD
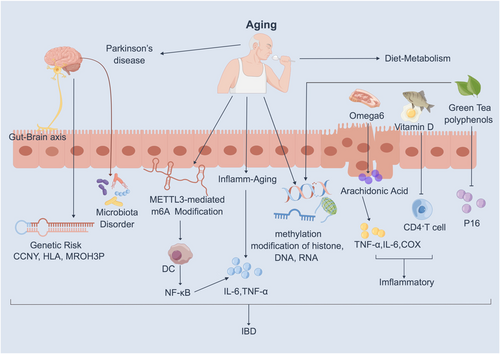
The gut–brain axis, which denotes interactions between the gut and the brain, has been a focal point in recent studies.46 Mounting evidence indicates that IBD contributes to the development of Parkinson's disease (PD). It is well established that PD is a neurodegenerative disorder affecting the aging brain, leading to a gradual decline in motor functions, including muscle rigidity, tremors, slowed movement, and impaired coordination. IBD and PD share common genetic risk factors/loci, such as MROH3P, HLA, CCNY, GUCY1A3, and BTNL2, underscoring the significance of the IBD–PD connection47, 48 (Figure 3). This implies that the role of gut inflammation will impact the onset of PD. Following an IBD diagnosis, understanding the peak age of onset for both diseases can aid in predicting and preventing PD.
The current studies have confirmed that IBD is associated with an increased expression of domain-containing protein 3 inflammasome. This association poses a heightened risk of neuroinflammatory disorders, such as Alzheimer's disease, and exacerbates cognitive impairment in older adults.49 Consequently, gut-selective immunosuppression may represent a safer therapeutic option for older patients. Additionally, the use of steroid medications for treating IBD can, in turn, worsen conditions such as hypertension, hyperglycemia, and congestive heart failure.50 Meanwhile, older patients encounter distinctive challenges, including recognizing the impact of comorbidities on the outcome of IBD treatment and understanding the crucial importance of establishing individualized treatment goals.
3.2 Age-related epigenetic modifications in IBD
Numerous studies have demonstrated a high correlation between the pathogenesis of inflammatory diseases and age-related epigenetic modifications.51-53 Aging augments METTL3-mediated methylation modification of m6A, thereby fostering DC activation and function and stimulating the NF-κB pathway (Figure 3). Moreover, the innate immune system expedites biological aging, a phenomenon termed “inflammatory aging,” wherein various inflammatory factors, such as TNF-α and IL-6, are activated, further exacerbating the development of IBD54 (Figure 3). Consequently, IBD patients with older age at onset exhibit a heightened risk of frailty compared to non-IBD elderly patients, and frailty is associated with an elevated risk of all-cause mortality. Therefore, targeting “inflammatory aging” linked to epigenetic alterations is crucial for diagnosing and treating patients with IBD.
A methylation group-wide association study involving 718 men and women aged between 25 and 72 years revealed a positive correlation between increased methylation of CpG islands and aging.55 These changes can be instigated by epigenetic modifications in the structure of DNA, RNA, and proteins (Figure 3). Upon analyzing typically aging colons, researchers observed a gradual loss of the methylated genome with age.56 It has been demonstrated that age-related methylation contributes to 70% of gene-specific methylation abnormalities in the colon.57 In conclusion, targeting methylation modification stands out as one of the potential therapeutic approaches for treating IIBD.
4 DIET METABOLISM REGULATES IBD VIA EPIGENETIC MODIFICATIONS
Research has demonstrated that dietary factors can modulate genomic expression and elicit modifications in host epigenetics, particularly among older individuals. A case in point is identifying vitamin D in meat and dairy products, exhibiting noteworthy in vitro antiproliferative, antibacterial, and anti-inflammatory properties. Vitamin D effectively impedes the activation of proinflammatory Th1 and Th17 cells and enhances T-cell functionality. Notably, the monoubiquitination of histone H2B has been found to diminish the activity of vitamin D receptors, subsequently promoting inflammation58 (Figure 3). Therefore, the administration of vitamin D supplements proves beneficial for IBD patients. Vitamin D influences the function of intestinal cells by binding to the vitamin D intracellular receptor, subsequently transcribing relevant genes.59 Consequently, a low vitamin D level is a biomarker and predictor of poor clinical outcomes in IBD patients. However, an increased intake of meat and dairy products, particularly those rich in omega-6 fatty acids, is associated with an elevated risk of IBD. Omega-6 is proinflammatory, and its derivative, arachidonic acid, acts as a precursor to primary proinflammatory mediators. This, in turn, activates inflammatory factors such as COX, lipoxygenase, IL-6, or TNF-α, ultimately triggering the development of IBD60 (Figure 3). Moreover, the active compounds in green tea, known as tea polyphenols (GTPs) or flavan-3-ols, possess the potential to potentially reverse aberrant methylation by modulating DNA methylation, histone modification, and microRNA epigenetic processes61 (Figure 3). High consumption of green tea and cruciferous vegetables demonstrated an inverse relationship with the methylation of trophectodermal markers (CDX2) and bone morphogenetic protein-2 (BMP-2), while this correlation was not observed for multiple tumor suppressor 1 (p16/INK4A), which is frequently hypermethylated in gastric cancers and serves as an essential senescence marker62 (Figure 3). Above all, initiating dietary control earlier proves beneficial in preventing IBD.
5 CONCLUSIONS AND FUTURE PERSPECTIVES
The pathogenesis of IBD has not been fully characterized, and its occurrence has been linked to various factors such as epigenetic alterations, diet, aging, and metabolism. These factors contribute to several barriers in the treatment and care of IBD. Aging and dietary metabolism induce alterations in epigenetic modifications in IBD, suggesting that these modifications play a role in diagnosing IBD.63 Therefore, targeting essential alterations in epigenetic modifications in IBD may represent a novel and efficient therapeutic strategy. However, it remains incompletely defined whether epigenetic modifications are altered as a result of the disease or if epigenetic modifications cause the disease. Furthermore, it is acknowledged that epigenetic modifications are dynamic and reversible, adding complexity to therapeutic strategies targeting these modifications.
Kaplan, Windsor, and others have projected that in the upcoming decade, individuals aged 65 and older with IBD will constitute over one-third of the total patient population, and elderly patients may experience a lower survival rate than their younger counterparts.64 Age-related differences in gut microbiota have been identified, with an increased prevalence of Akkermansia and a decreased abundance of Bacteroidaceae and Trichococcaceae.65 Consequently, there is an urgent need to establish personalized approaches for modifying the gut microbiota in treating elderly patients with IBD. Simultaneously, considering drugs utilized for treating aging-related diseases and delaying aging, such as rapamycin, metformin, spermidine, and aspirin, there is a possibility that these agents might be applicable in treating IBD as well. Considering the familial genetic predisposition of IBD, pursuing therapeutic targets grounded in alterations of epigenetic modifications may be the most effective strategy. While numerous studies have illustrated the crucial role of altered epigenetic modifications in various aging-associated diseases, understanding aging-associated epigenetic modifications in the context of IBD remains limited.
Immune metabolism plays a crucial role in regulating immunity and inflammation. As such, metabolic reprogramming of immune cells is a significant factor in inflammatory and anti-inflammatory responses. A notable example of this can be found in the role of IL-10, which controls macrophage function through metabolic reprogramming and aids in removing dysfunctional mitochondria. This process effectively inhibits proinflammatory responses, attenuating conditions such as IBD.66 Furthermore, impaired metabolism in epithelial cells has been shown to impact the regenerative capacity of the intestinal tract. Thus, it becomes clear that mitochondrial dysfunction in intestinal epithelial cells is a contributing mechanism to IBD pathogenesis. Mitochondrial dysfunction in intestinal epithelial cells is an early event in the pathogenesis of IBD, preceding inflammatory tissue abnormalities. However, it is yet to be determined whether these alterations are a cause or a consequence of the injury and subsequent inflammatory response. IBD can be considered an “energy deficiency disease” that affects the intestinal epithelium. Enhancing intestinal epithelial metabolism may strengthen barrier integrity and mucosal tolerance.67 A growing body of evidence suggests that targeting specific metabolic processes could serve as a strategy to mitigate inflammation in different scenarios.
In summary, although ample evidence suggests a connection between altered epigenetic modifications accompanying aging and diet metabolism and the development of IBD, the underlying mechanisms remain unclear. Additionally, while metabolic reprogramming during aging and dietary changes may play a pivotal role, extensive future research is necessary to validate this hypothesis.
AUTHOR CONTRIBUTIONS
Yanting Du: Writing—original draft (equal). Guo li: Project administration (equal). Yang Zhou: Project administration (equal). Min Zuo: Investigation (equal). Hu Wang: Project administration (equal). Yanan Liu: Project administration (equal); writing—review and editing (equal). Liquan Hong: Project administration (equal). All authors have read and approved the final manuscript.
ACKNOWLEDGMENTS
This project was supported by National Key Research and Development Programs of China (grant no. 2021YFA1102800), National Natural Science Foundation of China (grant nos. 92249304 and 32160161), the Hainan Provincial Key Research and Development Program (grant no. ZDYF2022SHFZ308), Hangzhou Normal University Graduate Scientific Research, and Innovation Promotion Project (grant no. 2022HSDYJSKY056).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
ETHICS STATEMENT
None.
Open Research
DATA AVAILABILITY STATEMENT
Not applicable.




