Emergence and detection of gastro-enteric virulence genes in non-pathogenic Vibrio cholerae: Implications for public health and water safety in African developmental regions
Abstract
Background
Water, an indispensable component of life, has in recent times been associated with the distribution/spread of gastro-enteric diseases especially as safe water is a concern in Southern, Eastern and Western African developmental regions.
Methods
This study evaluated the molecular fingerprints of potential gastro-enteric associated virulence genes of 58 somatic antigen non-agglutinating Vibrio cholerae 1/139 (SA-NAG-Vc-1/139) strains from domestic water sources. Strains were isolated and characterized applying both culture-based-microbiological techniques and molecular-fingerprinting of target-specific identification genes using simplex/multiplex PCR assay and ERIC-PCR fingerprints.
Results
Our study revealed diverse gene-based indices vis T3SS (29/58, 50.00%), T6SS (33/58, 56.9%), rtxA (37/58, 63.79%), rtxC (12/58, 20.69%), NAG-stn/sto (13/58, 22.41%), prtV (17/58, 29.31%), hlyA (41/58, 70.69%), nanH (40/58, 68.97%), mshA (44/58, 75.86%), chxA (37/58, 63.79%), hapA (17/58, 29.31%), ace (22/58, 37.93%), and cep (20/58, 34.48%) etc. Such results show that 53.45% (31/58) of isolates harboured more than three virulence associated genes while nanH, mshA, chxA, T6SS, T3SS, rtxA, hlyA, mshA, chxA, ace and cep fingerprints were detected predominantly with corresponding ilea-loop test positive strains. ERIC-PCR also showed multiple target specific repetitive intergenic consensus sequence regions ranging from 2 to 8.
Conclusions
This is an indication that the previously known non-pathogenic strains now harbour potential gastro-enteric virulence which may be controlled by more than one virulent gene dynamics. It also suggests a current potential shift in the virulence dynamics of V. cholerae strains recovered from the study area and a re-evaluated view of the previously non-pathogenic V. cholerae strains. Furthermore, the presence of such genes in SA-NAG-Vc-1/139 strains indicates a potential public health related concern. Although these detected potential gastro-enteric associated genes may be implicated in sporadic gastroenteritis, our result has re-emphasized their probable public health concern as they may be involved in both endemic and severe gastroenteritis cases, which suggests the need for water routine monitoring or surveillance.
Abbreviations
-
- AWD/C
-
- acute watery diarrhea and cholera
-
- BBEIAC
-
- bacteria based enterocyte infection associated conditions
-
- BEEIPREG
-
- biotechnology and emerging environmental infectious pathogens research group
-
- BIE
-
- bacterial infections of the enterocyte
-
- C/AWD
-
- cholera/acute watery diarrhea
-
- D
-
- dams
-
- DNTP
-
- deoxyribonucleoside triphosphate
-
- IC
-
- earth canals
-
- NAG
-
- non-agglutinating
-
- PCR
-
- polymerase chain reaction
-
- R
-
- rivers
-
- SA-NAG-T-1/139Vc
-
- somatic antigen non-agglutinating type-1/-139 V. cholerae
-
- TCBS
-
- thiosulphate citrate bile salts-sucrose
-
- WHO
-
- world health organization
-
- WWFEs
-
- wastewater final effluents
1 BACKGROUND
Water being an indispensable component of life has in recent times been associated with distribution and spread of gastro-enteric diseases especially as there is an increasing need for water supply due to lack of safe water in Southern, Eastern and Western African developmental region. Field investigators and the World Health Organisation (WHO) have recently reported that there is an increasing annual case fatality (greater than 4 million) of contaminated and unsafe water associated/related infection cases with annual mortality higher above 143,000 globally [1, 2]. In Southern, Eastern and Western African developmental region, such cases have been linked with the gastro-enteritis, cholera and acute watery diarrhea (AWD) potential pathogen (Vibrio cholerae) revealing the high clinical relevance of such strains. Major known globally implicated strain are the O1 and O139 V. cholerae strains with poor interest on the somatic antigen non-agglutinating strains (SA-NAG-1/139 V. cholerae). Such poor interest has also necessitated poor knowledge-base on the virulence potential of the strain as well as pathogenesis especially in domestic water sources. Suffice it to say that report from WHO since the last five-10 years shows these non-agglutinating O1/O139 V. cholerae (SANAG-1/139- Vc) such as O75 and O141 are potential outbreak associated strains, which reveal the need for an integrated surveillance study [3, 4]). Although pathogenesis and virulence amongst bacterial strains remains a mechanism of competition/survival in any environment, the products from such mechanism has produced deleterious effect on diverse environment especially when it's associated with man, animal or plant. Various potential pathogenic bacteria produce diverse gene products in multiple environments which affect and/or influence such environment positively or negatively. Those that affect the intestine, ilia and gastric enterocyte, cause conditions that are regarded as gastro-enteric diseases (gastro-enteritis). Other intestinal, ilia and gastric enterocyte defective conditions associated with bacteria are acute-watery-diarrhea AWD or diarrhea, cholera and dysentery. These bacteria based enterocyte infection associated conditions (BBEIAC) or bacterial infections of the enterocyte (BIE) have been reported by various investigators to involve diverse virulent genes with related names arising from their toxins, disease type and region of infection [5-8]. Notable example of such are ace (encoding accessory cholera enterotoxin), cep (encoding core-encoded pilin for adhesion), rtxA (repeat in toxin type (A), rtxC (repeat in toxin type (C), NAG-stn/sto (non-agglutinating heat stable enteric toxin), prtV (collagenase or novel metaloprotease gene), hlyA (hemolysin gene type (A), hlyB (a haemolysin secretin protein or gene), hlyC (a lactonising lipase gene) hapA (protease A gene), nanH (the neuraminidase gene: involved in sialic acid catabolism; it is in some cases involved in the sensitivity of host cells to cholera toxin), mshA (mannose-sensitive haemaglutinin type (A)and chxA (Cholix toxin or a ribosylating toxin). Other virulent markers may elicit proteases which are protein degrading enzymes (prtV), hemolysis (hlyA) which degrades haemoglobin etc [6, 9-11]. The multiple effects of these aforementioned effectors proteins/genes as well as other secretion system genes (T3SS and T6SS) results a disease condition which may be confirmed after cellular machineries of host is subverted and homeostasis is disrupted [12]. These genes present potential of eliciting pathogenic function amongst Vibrio cholerae strains even in the absence of core known pathogenic genes such as TCP, Tox and CTX genes [13].
Major DNA-based techniques proposed by most investigators for detection and targeted surveillance intervention of such enterocyte infecting bacteria cases are PCR specific gene detection, Enterobacterial Repetitive Intergenic Consensus sequence Polymerase Chain Reaction (ERICs-PCR), Repetitive Extragenic Palindromic Polymerase Chain Reaction (REP-PCR), Random Amplified Polymorphic DNA (RAPD) and Pulse Field Gel Electrophoresis (PFGE) [14-17]. Although the entero-pathogenesis associated with the cholera toxin (CTX in Vc) has been extensively studied, such virulo-relevant gene products are not common among SA-NAG-Vc-1/139. We are of the opinion that multiple virulo-relevant gene products may also be associated with gastro-enteric preponderance among the SA-NAG-Vc-1/139. Up till now, there has been dearth experimental evidence that associate these genetic fingerprints with such gastro-enteritis amongst environmental non agglutinating strains of V. cholerae. This study investigates/evaluates the molecular fingerprints of potential gastro-enteric associated virulence genes in non-pathogenic Vibrio cholerae strains isolated from domestic water sources in Southern, Eastern, and Western African developmental regions. This includes characterizing the genetic profiles of these strains using culture-based microbiological techniques and molecular fingerprinting assays, identifying virulence genes associated with gastro-enteric illnesses, and assessing their implications for public health. Ultimately, the study aims to inform evidence-based strategies for water quality management, disease prevention in resource-limited settings, address global health disparities and advance some sustainable development goals.
2 METHODS
Fifty-eight V. cholerae candidate-organisms for the study were isolated from water sources (domestic and surface water) commonly used within three local municipalities (Chris Hani, Amahlathi, and Lukhanji local municipalities) in Eastern Cape Province, South Africa and other municipalities in Nigeria, Botswana, Ethiopia, Uganda, Zimbabwe and Zambia. These V. cholerae isolates and our laboratory typed culture collections were gifted from our previous study at AEMREG laboratory South Africa, and shipped into our research group laboratory (Biotechnology and Emerging Environmental Infectious Pathogens Research Group (BEEIPREG)), Biotechnology unit, Department of Microbiology, Delta State University Abraka, Delta State, Nigeria for analysis. The strains were distinctly cultured onto thio-sulphate citrate bile salt agar (TCBSA) and further sero-grouped (after purity test) as somatic antigen non-agglutinating strains using the polyvalent antisera and molecular biology technique using Vc-O1F/Vc-O1R and Vc-O139F/Vc-O139R primer pairs sets as depicted in Table 1. Other in vitro culture-based gastro-virulent phenotypic determinants were employed such as Lipase detection (Ryan test), Protease detection (Skimmed milk agar test), Lecithinase detection (Egg York agar test) and Choleragen detection (Cholera red production test) and reported elsewhere [6, 7, 33, 34].
| Target gene | Primer name | Sequence | Expected Band size(bp) | Annealing Temp(°C) | Reference |
|---|---|---|---|---|---|
| 16S rRNA | VF169 | GGA TAA CC/TA TTG GAA ACG ATG | 617 | 53 | [18] |
| VR744 | CAT CTG AGT GTC AGT G/ATC TG | ||||
| OmpW | V. choleF | CACCAAGAAGGTGACTTTATTGTG | 304 | 64 | [19] |
| V. choler | GGTTTGTCGAATTAGCTTCACC | ||||
| Vc Serogrp | Vc-O1F | GTTTCACTGAACAGATGGG | 192 | 55 | [20] |
| Vc-O1R | GGTCATCTGTAAGTACAAC | ||||
| Vc-O139F | AGCCTCTTTATTACGGGTGG | 449 | 55 | [20] | |
| Vc-O139R | GTCAAACCCGATCGTAAAGG | ||||
| TSS | T3SS vcsV2-F | GGCTCACCAGCTGTTATGGT | 263 | 50 | [21] |
| T3SS vcsV2-R | CGTATTGCACAAGTAGCCGC | ||||
| T6SS vasH-F | TGTTGATGGGCGAGAGTCAC | 631 | 55 | [21] | |
| T6SS vasH-R | ACGTGTGTGGCAGATACCAG | ||||
| RTX | rtxA-F | CTG AAT ATG AGT GGGTGA CTT ACG | 418 | 55 | [22] |
| rtxA-R | GTG TAT TGT TCG ATA TCC GCTACG | ||||
| rtxC-F | CGA CGAAGA TCA TTG ACG AC | 263 | 55 | [22] | |
| rtxC-R | CAT CGT TAT GTG GTTGC | ||||
| Ace | ace F | TAAGGATGTGCTTATGATGGACACCC | 316 | 59 | [23] |
| ace R | CGTGATGAATAAAGATACTCATAGG | ||||
| Cep | cep F | GCTACATGTTTAGCTCACTG | 251 | 60 | [24] |
| cep R | TTTAGCCTTACGAATTAAGCC | ||||
| NanH | nanH F | CTTCCTCCAATACGGTTCTTGTCTCTTATGC | 314 | 55 | [25] |
| nanH R | TTCGGCTACCATCGGCAACTTGTATC | ||||
| chxA | chxA F | TGGTGAAGATTCTCCTGCAA | 421 | 49 | [26] |
| chxA R | CTTGGAGAAATGGATGCGCTG | ||||
| sto/stn | NAG-ST F | GAGAAACCTATTCATTGCA | 192 | 55 | [27] |
| NAG-ST R | GCAAGCTGGATTGCAAC | ||||
| PrtV | PrtV F | CATACTGAGATGCTCTACGAT | 864 | 55 | [28] |
| PrtV R | TTTCACCATGTTCGGGCGTGA | ||||
| HapA | hapA F | TCAACTACAACACCGCAGAC | 270 | 55 | [28] |
| hapA R | GACGACAATCCCAAGAAGAG | ||||
| MSHA | mshA F | CGCACAATGAGGTTCGCCAAG | 512 | 60 | [29] |
| mshA R | CCGAAAATTGACCGCCATTATC | ||||
| HLY | hlyAF | GTG CGT ATC AGC CTA GAT GA | 255 | 59 | [30] |
| hlyAR | CCA AGC TCA AAA CCT GAA A | ||||
| hlyBF | CAA GCC TTC GCC AAT AAC | 622 | 54 | [31] | |
| hlyBR | CCA CTT TTT TCC CTT CAC C | ||||
| hlyC F | AAATCCGCCACTTCTCTTC | 653 | 54 | [31] | |
| hlyC R | AATCAAAGCCACCAAGCC | ||||
| ERIC | Eric1 | ATGTAAGCTCCTGGGGATTCAC | Multiple | 48 | [32] |
| Eric2 | AAGTAAGTGACTGGGGTGAGCG | Multiple | 48 | [32] |
2.1 Hemolytic activity
The production of β-hemolysin by the isolates was conducted applying the method of [28] with few modifications. Briefly, blood agar based containing 13 g of agar was sterilized and a freshly pre-collected 5% rabbit RBC was added, mixed gently, dispensed in plates then allowed to polymerized. The pre-prepared agar plates were inoculated with pure overnight culture supernatant of isolates and incubated for 24 h. Positive report was recorded for isolates that show zones of clearance and haemolysis around isolates/colony grown on the surface of blood agar plates.
2.2 Gastro-enteritis production by isolates broth culture extract in test rabbit ligated ilea
Briefly, a total of twelve (12) apparently healthy rabbits were made to acclimatize in a cage and ten (10) were anaesthetized with 50% chloroform which put the rabbit to temporary sleep. The peritoneum was cut open and the small intestine was asceptically ligated in about 5 cm intervals for 8 regions/loops. The first loop was inoculated with 1 mL sterile water while the second was inoculated with 1 mL broth extract of positive control isolate. Loop three to eight were inoculated with 1 mL extract of each broth test isolates culture [35]. It is important to mention that each of the ligated rabbits ilea were inoculated with six to eight broth inoculum preparation of test isolates as described previously by related investigators.
2.3 DNA extraction
With few modifications from the previous methods of Magueri et al. [36], pre-prepared bacterial suspension was heated to boiling for genomic DNA extraction. Briefly, purified single gifted strains which were previously stored in aliquot of 2 mL graduated skirted Cryo preservation tubes ([Starlabs, Milton Keynes, UK] containing a 0.4 ml mixture of normal saline and glycerol [1∶1 or a 50% glycerol] in 0.6 ml of sterile de-ionised water) were allowed to thaw, subculture onto pre-prepared nutrient agar plate and incubated at 37°C for 24 h. Single culture colonies were harvested and suspended in sterile/nuclease-free tris ethylene diamine tetracetate (TE- 0.5 M) in an eppendorf microfuge tube (1.5 ml). Heat was applied on tubes to boiling or 100°C using Lasec dry heating block (www.lasec.co.za: Dri-Block DB-3D Techne) for 15 min, contents in microfuge-tubes were centrifuged at 15,000 r/min for 8 min in 4°C(Hermle Microfuge; Labortechnik GmbH Germany). Supernatant was collected as DNA template, stored at −20°C for PCR amplification experiments.
2.4 Enterobacterial molecular dynamics fingerprint of 1/139 SA-NAG-V. cholerae isolates
The patho-virulent diversity and associated gastro-enteric determining genes or fingerprints were detected applying simplex PCR detection, Multiplex PCR detection and Enterobacterial Repetitive Intergenic Consensus sequence PCR (ERICs-PCR) on serogrouped/serotyped isolates. The various entero-virulent and gastro-virulent associated genes applied are included in Table 1 below in addition to two sets of ERICs primers (ERIC1 5′-ATGTAAGCTCCTGGGGATTCAC-3′, ERIC2 5′-AAGTAAGTGACTGGGGTGAGCG-3′). All PCR reactions were conducted or carried out in 20 μL final reaction volumes, consisting of 1 μL of ERIC-primers, 1.0 μL of MgCl2, 10 μL of a GoTaq ℗G2 green master mix supplied in 2X Green GoTaq℗G2 reaction buffer containing pH: 8.5, dNTPs (400 μM each of dGTP, dTTP, dCTP and dATP), 3 mM MgCl2 and GoTaq℗G2 DNA polymerase at optimal concentration for efficient PCR amplification as specified by Promega Corporation USA (www.promega.com). Approximately 3.5 μL of dry-heat extracted DNA (as template) and 4.5 μL of nuclease-free water were simultaneously employed. The thermal cycling conditions for ERIC-PCR consisted time/temperature initial denaturation step at 6 min, 94°C, denaturation, annealing, and extension as follows: 35 cycles of (0.5 min, 94°C), (1 min, 48°C), (5 min, 72°C), and final extension (7 min, 72°C). Amplicons were stored at 4°C until post PCR procedures (electrophoresis detection and photography or documentation). Other genes detection thermal conditions are as reported in Table 1. A ten-microliter (10 μL) product of simplex/multiplex PCR and ERIC-PCR products were separated or electrophoresed using TakaRa Mupid-ONE (SNo: 073,031 and 073,032, Japan) after preparing a 1.5 g agarose powder in 100 ml TBE (Laboratorois Conda, Madrid, Spain). The 1.5% prepared agarose gel was stained with 2 μL ethidium bromide (1 μg/mL, Sigma-Aldrich, USA) submerged in a TBE buffer (pH 8.0) and loaded with PCR amplicons. An equivalent volume of both amplicons and commercially available PCRBIO of 100 bp to 10 kb pairs ladder (PCR Biosystems Ltd, London, UK) was filled onto the well that terminates each agarose gel casted as molecular marker before electrophoresis. The electrophoresed gel was thereafter transferred for visualization after electrophoresis of DNA amplified fingerprints and documented or photographed using UV transilluminator. Reference name and accession number of strains applied during study include O1 V. cholerae N16961 (acc. Num. NC_002505), O139 V. cholerae MO10 (acc. Num. NZ_AAKF00000000), O1 V. cholerae CIRS101 (acc. Num. NZ_ACVW00000000), O1 V. cholerae B33 (acc. Num. NZ_ACHZ00000000), and O1 V. cholerae MJ-1236 (acc. Num. NC_012668). These strains and primer pairs were previously designed/developed by NICD, South Africa for multiplex PCR. Other typed culture collections from our laboratory were also analyzed with reference strains to confirm gastro-enteric virulo-relevant associated genes. These reference strains and nuclease-free water were adopted as positive control and negative control respectively for PCR assay.
2.5 Evaluation of molecular fingerprints data
Procedural standardization, validation, amplified products and gel bands visualization data were normalized while fingerprints (Simplex/Multiplex and ERIC PCR) were scored manually, as “1” for a detected gene and “0” for an absent gene. The binary data were evaluated following the Jaccard coefficient of similarity and phylogeny dendrogram. The details of comparism were constructed using the Unweighted Pair Group Method with Arithmetic mean (UPGMA) [37]. The details of gel reports interpretation based on positive and negative detection were imported onto PAleontological Statistics 3.23 version (PAST3.23v) [38] for similarity and diversity studies. The dendrogram of the matrices was developed using neighbor-joining (NJ) [39] of the Euclidean similarity index (Equation 1), while The differentiating/diversity power was determined using the Simpson's coefficient of diversity (D) by using the formula D = 1{Σ[nj(nj1)]}/[N(N1)], where N is the number of strains tested, and nj denotes the number of strains belonging to the jth type which = 2–8 number of bands (of representative strains) produced by each isolate. Secondly, the numerical density of diverging strains in each clade. This is followed by a comparative resolution power of each fingerprinting as the ratio of number of clades for strains from its NJ clustering to the total number of isolates tested.
3 RESULTS
3.1 Isolation and identification of Vibrio cholera
Fifty-eight V. cholerae candidate-organisms were isolated from water sources (domestic and surface water) commonly used within three local municipalities (Chris Hani, Amahlathi, and Lukhanji local municipalities) in Eastern Cape Province, South Africa and other municipalities in Nigeria, Botswana, Ethiopia, Uganda, Zimbabwe and Zambia as reported from our previous studies. These strains were further characterized as somatic antigen non-agglutinating strains using the polyvalent antisera and molecular biology technique using Vc-O1F/Vc-O1R and Vc-O139F/Vc-O139R primer pairs sets as depicted in Table 1. Other in vitro culture-based gastro-virulent phenotypic determinants were employed such as Lipase detection (Ryan test), Protease detection (Skimmed milk agar test), Lecithinase detection (Egg York agar test) and Choleragen detection (Cholera red production test) and reported elsewhere (Igere et al., 2022a,b,c).
3.2 Haemolysis and in vivo production of gastro-enteritis by test isolates broth in ligated rabbit ilea
All tested isolates were negative to the sero-dynamic molecular study which confirm their non-somatic agglutinating reports. Amongst the 58 isolates tested, 27 (46.55%) showed positive haemolysin phenotype, as a clear transparent lytic zone around isolate indicates positive haemolysis whereas 21 (36.21%) showed positive gastro-enteritis production in in vivo ligated ilea loop test as a positive gastro-enteritis result was reported with the observation of an accumulation of fluid (as swollen intestine) in each ligated loop due to the production of adenylate cyclase and guanylate cyclase which were later converted to both cyclic 3,5-adenosinmonophosphate (AMP) and 3,5-guanosinmonophosphate (GMP) in an energy dependent reaction (Figure 1). As previously reported by related investigators, the produced cyclic AMP and GMP, further induce accumulation of sodium and chloride ion (a hydroscopic compound) and result absorption of water as well as passage of watery stool or diarrhea. This is an in vivo gastro- enteric infection induction in the rabbit ligated ilea as previously reported in investigators related studies [40, 41].
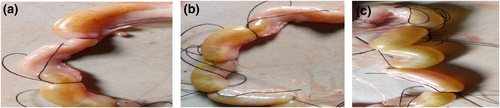
Showing in vivo production of gastro-enteritis by test isolates broth in ligated rabbit ilea. (a) Above shows three ligated ilea with two positive gastroenteritis induction and one negative result. (b) Shows four ligated ilea loop with three positive and one negative results. (c) Shows six ligated ilea with one of the ligated ilea loop of animals tested showing negative whereas five loops were positive for tested isolates indicating potential gastroenteric disease.
3.3 Virulent genotype of gastro-enterocyte infections
A number of gastro-enterocyte virulence associated genes were detected by multiplex-PCR genotyping and simplex PCR detection. These includes the type-VI secretion system (T6SS), type III secretion system (T3SS), which are membrane-embedded, spear/pin-pointed-like weapon that are involved in Gram-negative strain virulence, competition and may also be applicable beyond clinical cases. Other detected potential genes are rtxA, rtxC, NAG-stn/sto, prtV, hlyA, nanH, mshA, chxA, hapA, ace, and cep. A careful observation of the detected genes shows that the phenotypic expression of virulence is a mirror-image of the genotypic detection which represents the effector activity of multiple detected genes produced in an in vivo phenotypic virulent determination amongst strains. It may also imply that a single detected genotype does not control the virulence phenotype observed amongst the somatic antigen non-agglutinating V. cholerae type 1/139 (SA-NAG-Vc-1/139) (Figures 2-7).
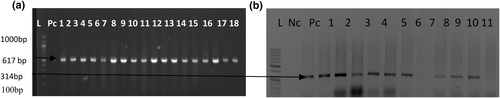
(a) Shows a photomicrogram of genus specific 16SrRNA gene simplex molecular fingerprints showing band size of the isolates at 617 base pair, while (b) Shows a photomicrogram of specie specific OmpW gene (304bp) simplex molecular fingerprints showing band size of the isolates.
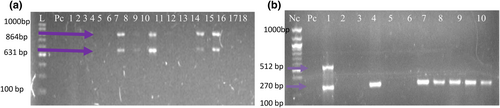
(a) Shows a photomicrogram of duplex PCR for PrtV and hlyB genes with band size for the isolates at 864 bp and 622 bp whereas (b) Show a photomicrogram of a duplex PCR amplicon for MSHA and hapA showing the band size of isolates at 512 bp and 270 bp.

(a) Shows the photomicrogram of simplex PCR for ChxA gene showing the band size for the isolates at 412 bp while (b) Show the photomicrogram of a duplex PCR for PrtV and ace genes showing the band size for tested isolates at 864 bp and 316 bp.
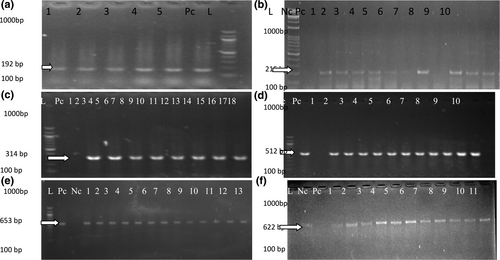
Shows the photomicrogram of PCR products revealing the band size of tested strains for Non-sto/stn, cep, NanH, mshA and hlyC, hlyB which are potential enterocyte associated virulent genes at expected band size of 192 bp, 215 bp, 314 bp, 512 bp, 653 bp and 622 bp.
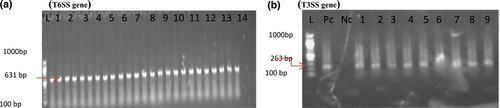
(a) Shows the photo of the T6SS gene (631 bp) in strain number 1–24, while (b) Represents photo of the T3SS gene detection (263 bp) in strain number one to nine.
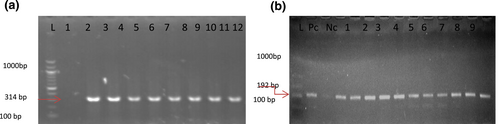
(a) Shows the photo of the NanH gene (314 bp) in strain number 1–24, while (b) Represents photo of the sto/stn gene detection (263 bp) in strain number one to nine.
4 DISCUSSION
The effector protein products released by most systemic and environment inhabiting organisms due to survival mechanism has been linked with diverse deleterious effect and diseases which has continued to affect greater global population at some instance. Such genes or proteins have been linked with diverse health related situation associated with man and environment, as it have been reported in various cases. This study reveals the molecular fingerprints of multiple potential gastro-enteric associated virulence associated genes amongst non-pathogenic Vibrio cholerae strains (SA-NAG-Vc-1/139) recovered/isolated from water sources in Southern, Eastern, and Western African developmental regions. It was observed from the study that all strains were negative to both the orf-O1 and orf-O139 genes detection using PCR and somatic antigen non-agglutinating polyvalent antisera (Figure 2a,b, Tables 2 and 3) which reveal them as NAG strains of V. cholerae and also presumptively labeling them as non-pathogenic. These strains were positive to the in vitro haemolysis test (27/58, 46.55%) which implies that they are producers of haemolysin. Accordingly, induction of gastro-enteritis (21/58, 36.21%) was also observed in isolates inoculation of rabbit ligated ilea using the experimental rabbit in vivo ligated ilea loop test (Figure 1). This implies that positive strains were potential producers of cholera/diarrhea, if expose to such environment. Similar gastro-enteric infection associated expression and character has been previously reported amongst the enterobacteriaceae members by diverse investigators [42-44]. In addition, such gastro-enteric infection associated phenotypes/expression are predominant amongst O1/O139 V. cholerae members. However in recent times, such expression have been reported amongst SA-NAG-Vc-1/139 (this study) and other environmental enteric strains/pathogens [4, 20, 45-50] in Cuba, America, Africa, China and other places in Bangladesh, India. This is a notable public health implication and indicates a potential concern to the public especially as the call on safe-water for all has become a global interest.
| Sites/Sources | Confirmed isolates (OmpW) | rfb-O1 | rfb-O139 | Ilea loop | Haemolysis | T3SS | T6SS | toxR | rtxA | rtxC |
|---|---|---|---|---|---|---|---|---|---|---|
| Wastewater treatment plants | 30 | 0 | 0 | 8 | 13 | 14 | 16 | 12 | 17 | 5 |
| River water | 16 | 0 | 0 | 7 | 9 | 8 | 9 | 7 | 12 | 5 |
| Household wastewater | 10 | 0 | 0 | 6 | 5 | 6 | 6 | 5 | 7 | 2 |
| Canal water | 2 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 1 | 0 |
| Total | 58 | Nil | Nil | 21 | 27 | 29 | 33 | 24 | 37 | 12 |
| Sites/Sources | NAG-stn/sto | hlyA | prtV | nanH | mshA | chxA | hapA | Ace | Cep | hlyB |
|---|---|---|---|---|---|---|---|---|---|---|
| Wastewater treatment plants | 5 | 20 | 6 | 23 | 24 | 21 | 8 | 9 | 8 | 11 |
| River water | 3 | 12 | 8 | 11 | 14 | 11 | 6 | 7 | 6 | 6 |
| Household wastewater | 3 | 8 | 3 | 5 | 6 | 5 | 3 | 5 | 4 | 2 |
| Canal water | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 0 |
| Total | 13 | 41 | 17 | 40 | 44 | 37 | 17 | 22 | 20 | 19 |
Apart from haemolysis and the in vivo induction of gastro-enteric infections potential amongst the reported strains, the following potential virulent genotypes of gastro-enteric infection relevance were also detected with details as follows: T3SS (29/58, 50.00%), T6SS (33/58, 56.9%), rtxA (37/58, 63.79%), rtxC (12/58, 20.69%), NAG-stn/sto (13/58, 22.41%), prtV (17/58, 29.31%), hlyA (41/58, 70.69%), nanH (40/58, 68.97%), mshA (44/58, 75.86%), chxA (37/58, 63.79%), hapA (17/58, 29.31%), ace (22/58, 37.93%), and cep (20/58, 34.48%) using simplex/multiplex PCR assay (Figures 3-7, Tables 2 and 3). The study revealed that 50.8% (31/61) of the isolates harboured more than three virulent associated genes with nanH, mshA, chxA, T6SS, T3SS, rtxA, ace and cep fingerprints being predominant amongst detected genes. According to various earlier global investigators reports, these aforementioned virulent genes have been found both in isolates from cholera outbreak cases and in clinical V. cholerae cases of diarrhea in hospitalized patients [46, 48, 50-52]. This implies that such non-pathogenic strains may be pathogenic and may also be implicated in potential outbreak cases which necessitate adroit surveillance in any related case study. It is also important to note that the detected genes have been affirmed to contribute in effective colonization of intestinal mucosa or rabbit ilia model causing diarrhea/cholera etc, that is as severe and similar to the clinical presentations associated with CT, Tox and TCP genes [49, 53] among O1/O139 V. cholerae. Similar observations have also been reported amongst environmental strains, as notable examples of such V. cholerae members are the non-agglutinating V. cholerae strains [54-57]. This implies that the SA-NAG-Vc-1/139 applied in this study is currently evolving with diverse virulo-relevant indices which may produce potential case severity in future outbreaks cases and necessitates evaluation of management strategies.
The observation of T3SS in this study further affirms the patho-virulent severity among strains recovered during the study. A similar report was previously recorded by related investigators affirming that T3SS and other virulent genes [58-60], among V. cholerae AM19226, NRT36S and N16961 strains were carriers and/or encode Vibrio Pathogenicity Island (VPI) and produce severe cholera as well as secretary diarrhea response in infected enterocytes. Suffice it to say that T3SS gene is secretion gene type which may be present in most Gram negative as well as Vibrio species. Furthermore, it has been previously demonstrated that the secretion system type three is a functional gene product amongst entero-pathogenic bacteria strains and may be responsible for intestinal colonization both in rabbit ilia and man [41, 53]. These gene members amongst other related virulent genes are present in most Gram-negative potential pathogens (V. cholerae) which suggest and affirm the gastro-virulent as well as entero-virulent relevance of these studied strains.
In addition, the type six secretion (T6SS) gene is the other secretary-based enterocyte associated virulent determinant detected during the study. It is shown to be widely distributed amongst Gram negative bacterial and the Vibrio species where it functions as a protein translocation/transportation machinery and inter-bacterial antagonism [58, 61-64]. It has resemblance with an inverted phage which has 14 proteins, complexed into three assembly which work interactively to transport protein via contractile mechanism [65, 66]. According to Fu et al. [67] and Chen et al. [58], the mechanism of virulence arising from T6SS genes has remained unknown however; there is high chance that it may be associated with virulence/pathogenesis of SA-NAG-Vc-1/139. Hence our observation of T6SS genes suggest the potential for pathogenesis/virulence as previously reported [58, 67].
There have also been various report of genotyped enteric borne pathogens both of clinical origin (diarrhea stool/faecal samples etc), food (sea food, animal products, vegetables, seeds etc) and environmental (contaminated water, rocks etc) sources using the ERIC-PCR and other PCR techniques [37]. These molecular tools are chosen especially as the pathogens are members of the broad family enterobacteriaceae. We employ these tools as an approach amongst emerging choleragenic Non-O1/Non-O139 to compare their intraclonal diversity/relatedness and the gastro-virulent potential of such strains with specific genetic mosaicism in the environment [5, 68-73]). From our study, there was an observed considerable genetic relatedness amongst the isolates from similar sources, a notable variation was found in the profile generated by the enterobacterial repetitive intergenic consensus sequence PCR (ERIC-PCR) and other PCR determination applied. There was an observable identical base pair banding pattern amongst isolates of similar serogroup and potential pathogenic members but those of null pathogenic potential had a different banding pattern from potential pathogenic strains with high discriminating index. Such discriminatory index was observed in some of the isolates which were previously described/identified as non-pathogenic. The necessity to emphasize on regular and periodic surveillance of water bodies as well as effluents is of epidemiological relevance.
The Enterobacterial Repetitive Intergenic Consensus sequence PCR (ERICs-PCR) detected regions divided the isolates into two clade where isolates number codes such as 35, 31, 39, 30, 45, 34, 433, 411, 181, 33,739, 38 harbors already know potential entero-virulent and gastro-virulent associated genes whereas, other isolates harbor emerging gastro-enterocyte/gastro-enteritis determinants. These specified strains (1/139 SA-NAG- V. cholerae) were further divided into two clades based on their gastro-enterocyte/gastro-enteritis determinants (Figure 8a). The observation was also repeated in the PCA Scatter diagram (Figure 8b) showing biplot and 96% eclipse of isolates, while isolates with number codes 30, 35 and 433 fell outside the eclipse region of relationship. This is a potential indication that gastro-enteritis is controlled by multiple gene function amongst strains.
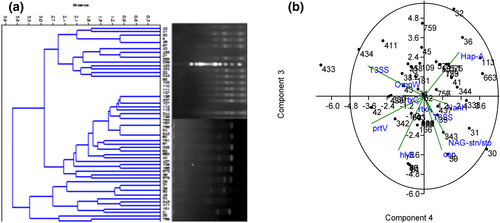
(a) Photomicrogram of ERIC-PCR-1, ERIC-PCR-2, and Neighbour joining clustered dendrogram generated by the past3.zip 3.14 version software which represents Euclidean similarity clade of gastro-enteritis producing isolates. Two related clustered clade of branching Isolates (b) PCA scatter diagram with Biplot, and 96% eclipse of the isolates with their various potential gastro-enteritis gene while Isolates 30, and 433 fell outside the 95% eclipse region of relationship.
Our study specific limitation include the geographical scope, sample type and/or size, in vitro genomic expression of the detected genes which are basic components of pathogenicity island for V. cholerae strains and non-detection of other gastro-virulence associated genes among bacterial members. The negative detection of some suggested genes as well as their potential implications for the study were not exhaustively determined by applied methods. It should be noted that the application of MLST would describe such concerns however MLST was not applied due to available resources during study. Furthermore, applied methods for gastro-enteritis was only affirmed using gastroenteritis related gene detection using PCR and rabbit ligated ilea tests. The above are limited area of potential concerns which are current ongoing studies aspect in our laboratory in a bid to affirm the state of the previously non-pathogenic nature of somatic antigen non-agglutinating V. cholerae type 1/139 (SA-NAG-Vc-1/139) and their evolutionary status in study area.
5 CONCLUSIONS
This study has described the production of gastro-enteritis virulence determinants of water origin or environmental V. cholerae strains and the associated emerging potential public health concerns. It has revealed the presence of diverse virulent genotypes including T3SS, T6SS, rtxA, rtxC, NAG-stn/sto, prtV, hlyA, nanH, mshA, chxA, hapA ace and cep. It has reaffirmed that the non-O1/non-O139 V. cholerae strains are potential etiological pathogen with the most recorded severe cholera and acute watery diarrheal disease epidemics which basically thrives in estuaries, coastal waters and environment. The strategy applied for the detection of enterocyte infecting potential pathogens will enhance potential enteric pathogens surveillance schemes and broaden our understanding in the development of Vibrio based vaccine for endemic, epidemic and pandemic strain (VVEEPS). Such cross-sectional and systematic assessment of the environmental waters remains an apt strategy for prevention and prediction of possible impending outbreaks.
AUTHOR CONTRIBUTION
Bright Esegbuyota Igere: Conceptualisation, methodology, investigation, data curation, analysis, interpretation, writing—original draft preparation, writing—review and editing.
ACKNOWLEDGMENTS
The author wishes to acknowledge the contributions of Prof Nwodo Uchechukwu Uche, Prof Igbinosa Etinosa Ogbomoede, Prof Emmanuel Erufoare Odjadjare, Prof Okoh Anthony Ifeanyi and a crowd of diverse related investigators on vibriology. It is hoped that in the near future the menace of cholera and other associated infectious cases would be adequately controlled by major research contributions. The author acknowledges the funds provided by African German Network of Excellence in Science 2022 (AGNES-2022) and Govan Mbeki Research and development centre. Supervisor-Linked Bursary-2017 (SLB-2017).
CONFLICT OF INTEREST STATEMENT
The author declares no competing interest.
ETHICS STATEMENT
Our University/institutional research ethical committee approved the study with number DOU86/2023. Isolates were environmental strains, while animal blood was collected as advised following required consent.
INFORMED CONSENT
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The datasets/information used for this study are available within the manuscript and the Figure S1.




