XRCC1, ABCB1, CYP3A5 and GSTP1 gene polymorphism associated with platinum-based drugs induced hematotoxicity in Chinese oesophageal cancer patients
Shuang Chen and Xiao Xiao have contributed equally to this work.
Abstract
Background
Hematotoxicity, including severe myelosuppression, is a common adverse drug reaction (ADR) during platinum-based treatment for oesophageal cancer (EC).
Purpose
The aim of this study was to identify single-nucleotide polymorphisms (SNPs) associated with platinum-induced hematotoxicity in patients with EC, as the relationship between SNPs and this ADR is incompletely demonstrated.
Methods
A total of 262 patients receiving platinum-based chemotherapy (cisplatin, nedaplatin, carboplatin and oxaliplatin) were enrolled in this study. Ten SNPs in eight genes were genotyped via multiplex polymerase chain reaction and sequenced to evaluate their relationship with severe myelosuppression and its subset of leukopenia and neutropenia.
Results
Multivariate logistic analysis of cisplatin cohort in severe leukopenia group showed an odds ratio (OR) of GG + GA versus AA in ABCB1 rs1045642 was 5.83 (95% confidence interval (CI) 1.63–20.83, p = 0.007, false discovery rate (FDR) = 0.028), while the OR of AA versus AG + GG in rs1128503 was 0.34 (95% CI 0.14–0.86, p = 0.022, FDR = 0.044). In nedaplatin cohort of neutropenia group, the OR of AA versus GG, AA + AG versus GG in GSTP1 rs1695 was 10.34 (95% CI 1.71–62.40, p = 0.011, FDR = 0.040) and 7.48 (95% CI 1.37–40.81, p = 0.020, FDR = 0.040) respectively. XRCC1 rs1799782 GG genotype in nedaplatin cohort of myelosuppression group and CYP3A5 rs776746 CT genotype in nedaplatin cohort of severe leukopenia group and platinum cohorts of all groups appeared to be risk factors with the p values less than 0.05, but FDR values were all greater than 0.05.
Conclusion
This study identified SNPs of XRCC1, ABCB1, CYP3A5 and GSTP1 related to hematotoxicity of platinum-based drugs, thereby providing a novel theoretical basis for the prediction and prevention of ADRs in platinum-based chemotherapy.
Abbreviations
-
- ABCB1
-
- ATP binding cassette subfamily B member 1
-
- ABCG2
-
- ATP binding cassette subfamily G member 2
-
- ADR
-
- adverse drug reaction
-
- BER
-
- basal excision repair
-
- BMI
-
- body mass index
-
- CTCAE
-
- Common Terminology Criteria for Adverse Events
-
- CYP3A5
-
- cytochrome P450 family 3 subfamily A member 5
-
- EC
-
- oesophageal cancer
-
- GSTP1
-
- glutathione S-transferase pi 1
-
- MTHFR
-
- methylenetetrahydrofolate reductase
-
- NCI-CTACE
-
- National Cancer Institute-Common Terminology Standards for Adverse Events
-
- P-pg
-
- P-glycoprotein
-
- SLC19A1
-
- solute carrier family 19 member 1
-
- SNP
-
- single nucleotide polymorphism
-
- TNM
-
- tumour–node–metastasis
-
- TP53
-
- tumour protein 53
-
- XRCC1
-
- x-ray repair cross complementing 1
1 INTRODUCTION
Oesophageal cancer (EC) is one of the most common cancers worldwide in terms of incidence 604,100 new cases and death 544,076 in 2020, and China is one of the top five countries with a high incidence of EC worldwide [1]. Most patients with EC are not diagnosed timely until the advanced stage, resulting in a poor prognosis. Chemotherapy is an effective means to delay the progression of EC patients [2]. Platinum-based drugs, which inhibit DNA synthesis, are widely used in anticancer treatment, but they frequently have side effects [3].
Myelosuppression is a potentially fatal adverse drug reaction (ADR) of chemotherapy, which is specifically manifested as leukopenia, neutropenia, haemoglobin, or thrombocytopaenia, and may ultimately lead to treatment interruption or failure [4]. Symptomatic treatment measures for the injection of growth factors have been proven effective for myelosuppressive clinical situations. But they also brought financial and physical burdens to patients [5]. Here we discovered Single-nucleotide polymorphisms (SNPs) are related to the ADRs of platinum-based chemotherapy. The SNPs of the following genes involve platinum-induced ADRs: I. Tumour protein 53 (TP53) initiates apoptosis when DNA damage repair failed. II. Methylenetetrahydrofolate reductase (MTHFR), solute carrier family 19 member 1 (SLC19A1) and x-ray repair cross complementing 1 (XRCC1) participate in DNA synthesis and repair. III. ATP binding cassette subfamily B member 1 (ABCB1), ATP binding cassette subfamily G member 2 (ABCG2), Cytochrome P450 family 3 subfamily A member 5 (CYP3A5) and glutathione S-transferase pi 1 (GSTP1) are involved in the process metabolism and detoxification in platinum-based drugs. Identifying specific biomarkers of severe myelosuppression is crucial for predicting ADRs.
The loss of TP53 function aggravates cisplatin resistance and triggers apoptosis when DNA damage could not be effectively repaired [6]. The CC genotype and altered amino acid sequence (Arg72Pro) of TP53 rs1042522 have been linked to increased incidence of grade III/IV neutropenia [7, 8]. XRCC1 is a pivotal DNA damage repair gene, and its inhibition enhances cisplatin cytotoxicity [9]. XRCC1 rs25487 and rs1799782 have been applied to evaluate the effectiveness of platinum-based chemotherapy [10]. Adequate intake of dietary folate has been reported to reduce the risk of EC [11]. In other DNA repair pathways involving folate transport, MTHFR rs1801133 decreases the incidence of thrombocytopaenia and gastrointestinal toxicity during chemotherapy [12]. SLC19A1 is a constitutive component of the main pathway for folic acid to enter the cell. The SLC19A1 rs1051266 C allele leads to lowered plasma folate levels in healthy populations [13].
The genes encoding drug-transport proteins and drug-metabolising enzymes are associated with the pharmacokinetics of chemotherapeutic drugs in vivo. ABCB1 and ABCG2 are multidrug transporters that participate in regulating the absorption of endogenous and exogenous substances [14]. Previous studies implied that ABCB1 rs1045642 and rs1128503 genotypes are associated with the occurrence of hematotoxicity of platinum-based drugs [15, 16]. Additionally, the dysfunctional variant ABCG2 rs2231142 has been found to suppress the metabolism of substrate drugs [17]. CYP3A5, the main cytochrome 450 enzyme expressed in the oesophagus, metabolises many potential carcinogenic compounds [18, 19]. CYP3A5 rs776746 allelic variation attenuates enzyme activity [20]. GSTP1 is an enzyme participating in the detoxification process, and its rs1695 G allele has been associated with a higher risk of myelosuppression than the A allele [21].
The in vivo dynamic processes of platinum-based drugs are closely linked to genes involved in tumour suppression, DNA repair and drug-metabolising enzymes. The SNPs of these genes may aid in predicting the individuals' ADRs for platinum-based chemotherapy.
2 MATERIALS AND METHODS
2.1 Study design and participant
Causality categories of ADRs and platinum-based drugs were assessed at first, which was based on the WHO-UMC system (http://www.who-umc.org/Graphics/24734, last accessed on 7 May 2023). A total of 262 patients with EC were sequentially recruited in the Cancer Hospital of Shantou University Medical College from November 2003 to October 2019. The inclusion criteria were as follows: (1) histologically confirmed EC, (2) >18 years old, (3) patients who received platinum-based chemotherapy (cisplatin: n = 119, nedaplatin: n = 142, carboplatin/oxaliplatin: n = 54) for at least 1 time; did not administer prophylactic G-CSF during the study period, and (4) no familial hereditary disease, liver or renal dysfunction (the concentration of serum creatinine is more than three times the upper limit of normal (350 μmol/L) or with the history of renal transplantation or dialysis; the concentration of serum transaminases more than three times the upper limit of normal 120 U/L) or with cirrhosis, cardiopulmonary dysfunction, pregnancy or lactation, severe cardiovascular disease and infection.
The patients' baseline data, including demographic information, medical history, biochemical measurements, medication information and ADRs, were obtained from the hospital database. Severe myelosuppression was defined as neutropenia, leukopenia, haemoglobin, or thrombocytopaenia (grade III/IV). Toxicity assessment was conducted in accordance with the Common Terminology Criteria for Adverse Events (CTCAE–Version 5.0).
Monitor changes in blood routine before and after chemotherapy. Before chemotherapy, an absolute neutrophil count ≥1.5 × 109/L, platelets ≥80 × 109/L, haemoglobin ≥80 g/L and white blood cell count ≥3.0 × 109/L and no bleeding tendency were observed. This study was approved by the Ethics Committee of the Cancer Hospital of Shantou University Medical College (Approval number: 201809) and the Ethics Committee of Guangzhou Red Cross Hospital (Approval number: 2020-109-02). This study was conducted in accordance with the Declaration of Helsinki. Each subject signed informed consent.
2.2 Sample collection and processing
Blood samples of the participants were collected and placed into EDTA anticoagulation tubes, temporarily stored at 4°C and then centrifuged at 3000 g/min for 10 min to separate plasma and blood cells. DNA was extracted using a TGuide blood genomic DNA kit (TIANGEN) and a TGuide M16 automatic nucleic acid extractor (TIANGEN). DNA concentration was detected by Nanodrop 2000 (Thermo Scientific) to ensure high quality of extracted genomic DNA.
2.3 Selection of SNPs and genotyping
The 10 SNP information of the candidate genes were obtained from the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/, last accessed on 7 May 2023) dbSNP database. The SNPs were selected on the basis of the following criteria: (1) reported to be associated with platinum-based chemotherapy, (2) potentially associated with the pharmacokinetics of drugs and tumour-related pathway and (3) minor allele frequency ≥0.05 in the Chinese population. The genotype was detected using multiplex polymerase chain reaction and sequencing in Sangon Biological Engineering Technology and Service Co. Ltd. After designing and synthesising primer pools containing SNP sites, amplification of the target SNP site sequence and Illumina-compatible sequencing library preparation were finished by PCR. The data from the HiSeq XTen sequence system (Illumina) were quality-controlled and then compared to the reference genome using BWA (Version 0.7.13-r1126). The genotypes of target sites were calculated by the Perl script.
2.4 Statistical analysis
SPSS software 25.0 was utilised to analysed data. Continuous variables were expressed as mean ± standard deviation. The normality of the distribution of the continuous variables was checked by the Shapiro–Wilk test and analysed using an independent sample t-test. Categorical variables were expressed as n (%) and a chi-square test was used for analysis. All participants' genotype and the expected genotype were tested and the Hardy–Weinberg equilibrium of candidate SNPs in the participants with EC was evaluated. For the analysis of SNPs and ADRs, the participants were categorised into different groups in accordance with the platinum-based drugs and the occurrence of ADRs. Single factor analysis was conducted, and covariates such as age, gender, BMI, TNM stage, outcome of histology, history of tobacco smoking and alcohol drinking, diabetes and radiotherapy were included in the logistic regression model to assess the relationship between ADRs and the 10 candidate SNPs. The Odds ratio (OR), 95% confidence interval (CI) and p value were calculated. False discovery rate (FDR) was calculated using the Benjamini–Hochberg method on SAS 9.4. The OR with a p value less than 0.05 and the FDR value less than 0.05 was considered significant. The forest plot was mapped by GraphPad prism 7.00.
3 RESULTS
3.1 Demographic characteristics of the study population
The causality categories of 3 ADRs and platinum-based drugs were all ‘possible’. A total of 262 EC participants who met the inclusion criteria were enrolled during the study period (Table 1). The study participants included 206 males and 56 females, with an average age of 60.94 ± 7.04 years and BMI of 20.43 ± 3.06 kg/m2. Among them, 167 (63.74%) had a history of tobacco smoking, 95 (36.26%) had a history of alcohol drinking, 31 (11.83%) had hypertension, 14 (5.34%) had diabetes and 129 (49.24%) received radiotherapy. In addition, the patients were clinically grouped on the basis of tumour–node–metastasis (TNM) tumour stage, with 28 (10.69%), 132 (50.38%) and 102 (38.93%) belonged to TNM stages I/II, III and IV, respectively. The participants' pathological types were as follows: 57 cases (21.76%) were adenocarcinoma/others and 205 (78.24%) were squamous carcinoma.
| Characteristics | Cisplatin | Nedaplatin | Platinum drugs |
|---|---|---|---|
| n = 119 (%) | n = 142 (%) | n = 262 (%) | |
| Age (years) | 60.18 ± 6.88 | 61.13 ± 7.15 | 60.94 ± 7.04 |
| Gender | |||
| Male | 94 (78.99) | 107 (73.35) | 206 (78.63) |
| Female | 25 (21.01) | 35 (24.65) | 56 (21.37) |
| BMI (kg/m2) | 20.05 ± 3.01 | 20.34 ± 3.27 | 20.43 ± 3.06 |
| Tobacco smoking | |||
| Never/quit | 42 (35.29) | 56 (39.44) | 95 (36.26) |
| Ever | 77 (64.71) | 86 (60.56) | 167 (63.74) |
| Alcohol drinking | |||
| Never/quit | 72 (60.50) | 94 (66.20) | 167 (63.74) |
| Ever | 47 (39.50) | 48 (33.80) | 95 (36.26) |
| Hypertension | |||
| Yes | 16 (13.45) | 14 (9.86) | 31 (11.83) |
| No | 103 (86.55) | 128 (90.14) | 231 (88.17) |
| Diabetes | |||
| Yes | 4 (3.36) | 5 (3.52) | 14 (5.34) |
| No | 115 (96.64) | 137 (96.48) | 248 (94.66) |
| Radiotherapy | |||
| Yes | 57 (47.90) | 84 (59.15) | 129 (49.24) |
| No | 62 (52.10) | 58 (40.85) | 133 (50.76) |
| Tumour–node–metastasis stage | |||
| I/II | 20 (16.81) | 12 (8.45) | 28 (10.69) |
| III | 56 (47.06) | 74 (52.11) | 132 (50.38) |
| III | 43 (36.13) | 56 (39.44) | 102 (38.93) |
| Histology | |||
| Adenocarcinoma/other | 16 (13.45) | 10 (7.04) | 57 (21.76) |
| Squamous carcinoma | 103 (86.55) | 132 (92.96) | 205 (78.24) |
| Myelosuppressiona | |||
| Never | 65 (54.62) | 104 (73.24) | 167 (63.74) |
| Ever | 54 (45.38) | 38 (26.76) | 95 (36.26) |
| Leukopeniaa | |||
| Never | 78 (65.55) | 111 (78.17) | 194 (74.05) |
| Ever | 41 (34.45) | 31 (21.83) | 68 (25.95) |
| Neutropeniaa | |||
| Never | 80 (67.23) | 118 (83.10) | 200 (76.34) |
| Ever | 39 (32.77) | 24 (16.90) | 62 (23.66) |
| Thrombocytopaeniaa | |||
| Never | 113 (94.96) | 128 (90.14) | 241 (91.98) |
| Ever | 6 (5.04) | 14 (9.86) | 21 (8.02) |
| Anaemiaa | |||
| Never | 107 (89.92) | 139 (97.89) | 246 (93.89) |
| Ever | 12 (10.08) | 3 (2.11) | 16 (6.11) |
- a Myelosuppression, leukopenia, neutropenia, thrombocytopaenia and anaemia ADRs' definition standard is to reach the grade III/IV.
The results displayed that 36.26% of patients receiving platinum-based chemotherapy had severe myelosuppression (grade III/IV). The incidence of severe myelosuppression in the cisplatin and nedaplatin treatment cohorts was 45.38% and 26.76%, respectively. The haematological toxicity was primarily characterised by leukopenia and neutropenia. Ninety-five participants were diagnosed with severe myelosuppression including 68 cases (25.95%) of severe leukopenia (grade III/IV), 62 cases (23.66%) of severe neutropenia (grade III/IV), 21 cases (8.02%) of thrombocytopaenia and 16 cases (6.11%) of anaemia.
3.2 Relation between baseline variables and ADRs of hematotoxicity
To examine the correlation of ADRs with baseline parameters, we performed independent sample t-test (continuous) and chi-square test (categories) (Table 2). The anti-cancer drugs were divided into three cohorts: cohort I, cisplatin; cohort II, nedaplatin; and cohort III, platinum-based drugs (cisplatin, nedaplatin, carboplatin and oxaliplatin). Then, the ADRs of hematotoxicity (grade III/IV) were divided into three classifications (severe myelosuppression, severe leukopenia and severe neutropenia) and analysed together with the baseline demographic characteristics. Among the baseline demographic variables, age, gender, BMI, TNM staging, diabetes, history of alcohol drinking and outcome of histology significantly displayed correlation with haematological toxicity in at least one of the drug cohorts. However, the history of tobacco smoking, hypertension and radiotherapy did not show any significant differences between the ADR and non-ADR groups.
| Characteristics | p* | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cisplatin | Nedaplatin | Platinum-based drugs | |||||||||||||||||
| Myelosuppression | Leukopenia | Neutropenia | Myelosuppression | Leukopenia | Neutropenia | Myelosuppression | Leukopenia | Neutropenia | |||||||||||
| ADR n = 54 | n-ADR n = 65 | ADR n = 41 | n-ADR n = 78 | ADR n = 39 | n-ADR n = 80 | ADR n = 38 | n-ADR n = 104 | ADR n = 31 | n-ADR n = 111 | ADR n = 24 | n-ADR n = 118 | ADR n = 95 | n-ADR n = 167 | ADR n = 68 | n-ADR n = 194 | ADR n = 62 | n-ADR n = 200 | ||
| Age (years) | 0.071 | 0.222 | 0.156 | 0.198 | 0.676 | 0.049 | 0.364 | 0.561 | 0.128 | ||||||||||
| Gender | Male | 0.035 | 0.011 | 0.068 | 0.247 | 0.113 | 0.573 | 0.074 | 0.004 | 0.092 | |||||||||
| Female | |||||||||||||||||||
| BMI (kg/m2) | 0.048 | 0.041 | 0.017 | 0.416 | 0.560 | 0.150 | 0.349 | 0.091 | 0.011 | ||||||||||
| Tobacco smoking | Never/quit | 0.129 | 0.068 | 0.084 | 0.435 | 0.748 | 0.482 | 0.224 | 0.117 | 0.095 | |||||||||
| Ever | |||||||||||||||||||
| Alcohol drinking | Never/quit | 0.103 | 0.040 | 0.174 | 0.388 | 0.279 | 0.675 | 0.699 | 0.627 | 0.654 | |||||||||
| Ever | |||||||||||||||||||
| Hypertension | Yes | 0.690 | 0.400 | 0.315 | 0.425 | 0.506 | 0.783 | 0.484 | 0.197 | 0.765 | |||||||||
| No | |||||||||||||||||||
| Diabetes | Yes | 0.040 | 0.118 | 0.102 | 0.610 | 0.300 | 0.851 | 0.272 | 0.392 | 0.657 | |||||||||
| No | |||||||||||||||||||
| Radiotherapy | Yes | 0.291 | 0.527 | 0.295 | 0.175 | 0.054 | 0.202 | 0.476 | 0.321 | 0.463 | |||||||||
| No | |||||||||||||||||||
| TNM stage | I/II | ||||||||||||||||||
| III | 0.042 | 0.131 | 0.304 | 0.142 | 0.208 | 0.120 | 0.030 | 0.312 | 0.325 | ||||||||||
| IV | |||||||||||||||||||
| Histology | Adenocarcinoma/other | 0.348 | 0.783 | 0.889 | 0.023 | 0.149 | 0.004 | 0.603 | 0.195 | 0.600 | |||||||||
| Squamous carcinoma | |||||||||||||||||||
- Note: Age and BMI was analyzed with independent sample t test; gender, TNM stage, history of tobacco smoking, alcohol drinking, diabetes and radiotherapy, outcome of histology were analyzed with chi-square test.
- Abbreviations: ADR, adverse drug reaction; BMI, body mass index; n-ADR, no adverse drug reaction; TNM, tumour–node–metastasis.
- *The significance of difference between ADR and n-ADR groups; Bold number: p values that less than 0.05.
In the gender analysis, the proportion of female in the ADR group was relatively higher than that in the non-ADR group (Table S1). In cohort I, females accounted for 29.63% (n = 16) of the patients in the ADR group with severe myelosuppression, while only 13.85% (n = 9) were in the non-ADR group (p = 0.035). Regarding the assessment of severe leukopenia, females accounted for 34.15% (n = 14) in the ADR group and 14.10% (n = 11) in the non-ADR group (p = 0.011). Similarly, in cohort III, 33.82% (n = 23) and 17.01% (n = 33) were female in the ADR and non-ADR groups, respectively (p = 0.004). In cohort I, the patients with low BMI were found to be more susceptible to hematotoxicity.
3.3 Association of candidate SNPs with platinum-based drugs induced severe myelosuppression
A total of 10 SNPs of 8 genes were investigated for their relationship with the hematotoxicity of platinum-based chemotherapy in EC patients (Table 3). This study defined the participants with severe myelosuppression (grade III/IV) as the experimental group, while those without or with mild (grade I/II) myelosuppression were regarded as the control group. Multivariate logistic regression analysis was performed using age, gender, BMI, history of tobacco smoking and alcohol drinking, diabetes, radiotherapy, TNM stage, and outcome of histology as covariates to examine the association between severe myelosuppression and the 10 SNPs (Figure 1 and Table S2). Here the distribution of genotypes was consistent with the Hardy–Weinberg equilibrium.
| Gene | Function | Single nucleotide polymorphism | Alleles | Chr |
|---|---|---|---|---|
| TP53 | TP53 pathway | rs10452522 | C > G | chr17:7676154 |
| XRCC1 | DNA repair pathway | rs25487 | C > T | chr19:43551574 |
| XRCC1 | DNA repair pathway | rs1799782 | G > A | chr19:43553422 |
| MTHFR | Folic acid metabolism pathway | rs1801133 | G > A | chr1:11796321 |
| SLC19A1 | Folic acid metabolism pathway | rs1051266 | T > C | chr21:45537880 |
| ABCB1 | Transporters | rs1045642 | G > A | chr7:87509329 |
| ABCB1 | Transporters | rs1128503 | A > G | chr7:87550285 |
| ABCG2 | Transporters | rs2231142 | G > T | chr4:88131171 |
| CYP3A5 | P450 enzymes | rs776746 | C > T | chr7:99672916 |
| GSTP1 | Non-P450 enzymes | rs1695 | A > G | chr11:67585218 |
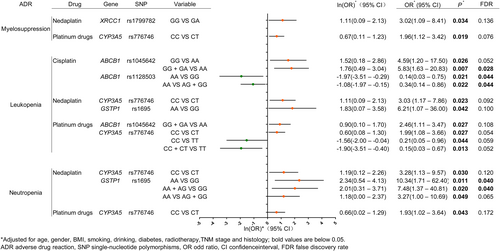
Correlation between candidate SNPs and severe ADRs. The effects of genes and SNPs on ADR are shown in the figure. Every ADR was analysed after classifying different platinum-based drugs, corresponding genes and SNPs, with further comparisons of specific genetic types. The forest plot was based on the Napierian Logarithm of the odds ratio value ln(OR), with the null value of 0, and the null line was defined as a vertical line of x = 0. The transverse lines lying on the left of the null line without intersection indicate that the mutation of the candidate SNPs reduces the occurrence of adverse events (myelosuppression, leukopenia, and neutropenia), and the effect amount ln(OR) is marked with green points. The transverse lines lying on the right of the null line without intersection indicate that the mutation of the candidate SNPs increases the occurrence of adverse events (myelosuppression, leukopenia, and neutropenia), and the effect amount ln(OR) is marked with orange points. ADR, adverse drug reaction; CI, confidence interval; FDR, false discovery rate; OR, odd ratio SNP, single nucleotide polymorphism.
Multivariate logistic regression analysis revealed that in cohort I, the 10 SNPs in eight candidate genes did not obviously affect the occurrence of severe myelosuppression (p > 0.05). In cohort II, we noticed that the XRCC1 rs1799182 GA genotype was associated with an increased incidence of severe myelosuppression compared with the GG genotype (OR = 3.02, 95% CI = 1.09–8.41, p = 0.034, FDR = 0.136). In cohort III, the CYP3A5 rs776746 CT genotype was a slight risk factor for severe myelosuppression (OR = 1.96, 95% CI = 1.12–3.42, p = 0.019, FDR = 0.076).
3.4 Association of candidate SNPs with severe leukopenia and neutropenia caused by platinum-based chemotherapy
The association between candidate SNPs and severe leukopenia or neutropenia (grade III/IV) was further analysed to determine whether the candidate SNPs affected the platinum-based drug-induced severe hematotoxicity. The patients with severe leukopenia or neutropenia (grade III/IV) were defined as the ADR group, while those without or with mild (grade I/II) leukopenia or neutropenia were regarded as the non-ADR group.
In cohort I, the analysis revealed that two SNPs, ABCB1 rs1045642 and rs1128503, were associated with severe leukopenia (Figure 1 and Table S3). In the models of ABCB1 rs1045642 GG versus AA (OR = 4.59, 95% CI = 1.20–17.50, p = 0.026, FDR = 0.052) and GG + GA versus AA, the AA genotype was a risk factor for severe leukopenia (OR = 5.83, 95% CI = 1.63–20.83, p = 0.007, FDR = 0.028). Moreover, the multivariate logistic regression analysis of ABCB1 rs1128503 demonstrated both the AA versus GG and AA versus AG + GG models, the double mutant GG genotype reduced the risk of severe leukopenia (OR = 0.14, 95% CI = 0.03–0.75, p = 0.023, FDR = 0.044; OR = 0.34, 95% CI = 0.14–0.86, p = 0.022, FDR = 0.044), respectively. This indicates that the G allele is associated with a lower risk of severe leukopenia compared with the A allele.
In cohort II, CYP3A5 rs776746 CT and GSTP1 rs1695 GG showed a correlation with severe leukopenia (Figure 1 and Table S3). The CYP3A5 rs776746 CT genotype was identified as a risk factor for severe leukopenia in comparison to the CC genotype (OR = 3.03, 95% CI = 1.17–7.86, p = 0.023, FDR = 0.092). Additionally, the GSTP1 rs1695 GG genotype was found to be a weak risk factor for severe leukopenia after adjusting the confounding factors (OR = 6.21, 95% CI = 1.07–36.00, p = 0.042, FDR = 0.100).
In cohort III, ABCB1 rs1045642 and CYP3A5 rs776746 were related to severe leukopenia (Figure 1 and Table S3). The ABCB1 rs1045642 with the G allele brought less risk of severe leukopenia than that with the A allele (GG + GA vs. AA: OR = 2.46, 95% CI = 1.11–5.47, p = 0.027, FDR = 0.108). For CYP3A5 rs776746, various mutant genotypes showed different trends with severe leukopenia. In contrast to the wild CC genotype, the CT genotype was identified as a risk factor for severe leukopenia (OR = 1.99, 95% CI = 1.08–3.66, p = 0.027, FDR = 0.054). Interestingly, the TT genotype seemed to be a protective factor against severe leukopenia (OR = 0.21, 95% CI = 0.05–0.96, p = 0.044, FDR = 0.059) and the T allele was proven to be a protective factor compared to the C allele in the CC + CT versus TT model (OR = 0.15, 95% CI = 0.03–0.67, p = 0.013, FDR = 0.052).
For severe neutropenia, none of the 10 SNPs showed any notable effect after cisplatin treatment (Figure 1 and Table S4). Similar to the trend observed for severe leukopenia in cohort II, the CYP3A5 rs776746 CT genotype was also a risk factor for severe neutropenia (OR = 3.28, 95% CI = 1.13–9.57, p = 0.030, FDR = 0.120). However, after correcting the confounding factors for GSTP1 rs1695, AA versus GG (OR = 10.34, 95% CI = 1.71–62.40, p = 0.011, FDR = 0.040), AA + AG versus GG (OR = 7.48, 95% CI = 1.37–40.81, p = 0.020, FDR = 0.040) and AA versus AG + GG (OR = 3.27, 95% CI = 1.00–10.69, p = 0.049, FDR = 0.065) indicated that the GG genotype was founded to be a risk factor for severe neutropenia. In cohort III, the CT genotype for CYP3A5 rs776746 was associated with a higher risk of severe neukopenia than the CC genotype (OR = 1.93, 95% CI = 1.02–3.64, p = 0.04, FDR = 0.172, Figure 1).
4 DISCUSSION
In this study, we analysed 262 blood samples and clinical data of patients with EC to investigate the relationship between 10 candidate SNPs and platinum-based chemotherapy-induced hematotoxicity. Our results verified that SNPs CYP3A5 rs776746 and GSTP1 rs1695 were linked to chemotherapy-induced hematotoxicity. Furthermore, we identified novel SNPs (XRCC1 rs1799782 and ABCB1 rs1045642 and rs1128503) that were related to hematotoxicity triggered by platinum-based chemotherapy. Most of them were in association with severe neutropenia and leukopenia. Above all, this finding supports the relationship between platinum-based drugs induced hematotoxicity and SNPs of XRCC1, ABCB1, CYP3A5, GSTP1, which have been shown to be potential predictors of ADRs in this study (p < 0.05). These genes are involved in the regulation of DNA repair function and drug metabolism enzyme activity. XRCC1 repairs damaged DNA by Ber. ABCB1, CYP3A5 and GSTP1 are involved in the process of metabolism and detoxification of platinum-based drugs (Figure 2).
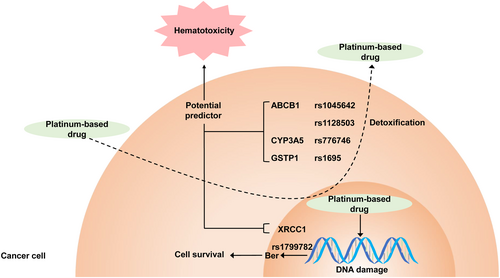
Relationship between platinum-based drugs induced hematotoxicity and SNPs of XRCC1, ABCB1, CYP3A5, GSTP1. Our study illustrates the single nucleotide polymorphisms of XRCC1, ABCB1, CYP3A5 and GSTP1 are related to the hematotoxicity during patients receiving platinum-based chemotherapy. These genes are involved in the regulation of DNA repair function and drug metabolism enzyme activity. XRCC1 repairs damaged DNA by Ber. ABCB1, CYP3A5 and GSTP1 are involved in the process of metabolism and detoxification of platinum-based drugs.
XRCC1 is a crucial protein involved in repairing single-strand breaks and base excision caused by ionising radiation and alkylating agents [22]. XRCC1 polymorphism disrupts its interaction with other enzymes, thereby impeding the DNA repair and leading to genetic instability and carcinogenesis [23]. Previous hints suggested that SNP rs1799782 (Arg194Trp) affects the DNA repair activity, but more research is needed to confirm its exact impact on gene function. The XRCC1 rs1799782 genotype has been proven to be a risk factor for liver cancer, EC and colon cancer [24-26] and a predictor of the response to platinum-based drugs on the basis of its A allele response [10]. In our study, the XRCC1 rs1799782 GA genotype was found to be a risk factor for severe myelosuppression compared with the GG genotype in nedaplatin chemotherapy (OR = 3.02, 95% CI = 1.09–8.41, p = 0.032). This study was the first to uncover the association between XRCC1 rs1799782 and severe myelosuppression. Another SNP, rs25487, has been associated with an increased risk of EC [27]. In our research, no obvious relationship was observed between rs25487 and the hematotoxicity of platinum-based drugs.
The ABC transporter superfamily reduces intracellular drug concentration by mediating intracellular drug efflux and causing drug resistance. P-glycoprotein, encoded by ABCB1, exhibits a wide substrate specificity with efflux-pump function and plays a significant role in multi-drug resistance in cancer [28]. A synonymous single nucleotide transformation of ABCB1 rs1045642 (G > A) attenuates the expression of P-gp in vivo [29], thereby potentially altering the P-gp protein and leading to differences in the efficacy and toxicity of chemotherapeutic drugs [30]. In fact, P-gp-positive L1210 cells exhibit reduced sensitivity to cisplatin [31]. In addition, alterations in cell regulatory pathways, such as protein expression, glycosylation and phosphorylation, have been demonstrated in cells overexpressing P-gp, which may consequently induce changes in cell sensitivity to substances that are not P-gp substrates or modulators [32]. Current studies have shown that ABCB1 rs1045642 and rs1128503 are associated with the prognosis and toxicity of chemotherapy. The rs1045642 A allele and rs1128503 A allele in ABCB1 have been identified as risk factors for neutropenia and anaemia, respectively [15, 33]. Here, we revealed the ABCB1 rs1045642 AA genotype was a risk factor for severe leukopenia, whereas the ABCB1 rs1128503 GG genotype was a protective factor during cisplatin chemotherapy. In conclusion, ABCB1 rs1045642/rs1128503 are closely related with severe leukopenia in platinum-based chemotherapy.
CYP3A5 is an important metabolic enzyme mainly expressed in the oesophagus, and its SNPs affect the enzyme activity and increase the risk of EC [20]. Studies have reported that CYP3A5 is expressed at higher levels in oesophageal squamous cell carcinoma tissue than in normal tissue [34]. In a study of platinum-based drug treatment for urothelial carcinoma, the CYP3A rs776746 T allele was found to be a significant gastrointestinal protective factor associated with reduced progression-free survival [35]. The CYP3A5 rs776746 TC genotype increases the risk of neutropenia in patients with breast cancer treated with docetaxel, epirubicin and cyclophosphamide [36]. Another study discovered that the CYP3A rs776746 C allele was a risk factor for neutropenia after docetaxel, doxorubicin and cyclophosphamide treatment [37]. In our study, we confirmed that the CYP3A5 rs77674 TC genotype was a risk factor for severe neutropenia and leukopenia, whereas the CYP3A5 rs77674 TT genotype was a protective factor for severe leukopenia patients receiving platinum-based drugs. These findings add to the growing body of evidence supporting that CYP3A5 genotyping can be useful in predicting chemotherapy-induced hematotoxicity and developing personalised treatment plans for cancer patients.
GSTP1, a II -phase detoxification enzyme located in 11q13, is mainly expressed in the oesophagus and thyroid [38]. GSTP1 rs1695 Ile105Val (A > G) leads to the spatial change in the GSTP1 substrate binding site by altering the amino acid sequence, thereby significantly changing the enzyme activity and inducing the detoxification of carcinogens, thus affecting the level of DNA damage [39]. GSTP1 rs1695, especially the AG/GG genotype, is a risk factor for susceptibility of various tumours, such as breast cancer [40]. Meanwhile, GSTP1 rs1695 also affects the risk of EC in the smoking population [41]. Regarding the hematotoxicity of platinum-based drugs (cisplatin, carboplatin and oxaliplatin), a meta-analysis indicated that GSTP1 rs1695 reduces neutrophils and the G allele is a risk factor for neutropenia [21]. In our study of nedaplatin chemotherapy, the GSTP1 rs1695 GG genotype was identified as a serious risk factor for leukopenia and neutropenia, consistent with previous research indicating the association of the G allele with neutropenia risk.
Genetic polymorphisms have been shown to be related to hematotoxicity in the group patients treated with platinum-based drugs. However, some SNP-cohorts of specific drugs did not show this relation, possibly due to the small sample size of each cohort. Despite this, the results of our study were still meaningful, as five SNPs showed a significant relationship between the SNPs and hematotoxicity, including two SNPs that confirmed previous reports and three SNPs newly discovered. Therefore, these data are credible and these genes are useful for the development of individualised chemotherapy as a part of guidance for the prediction and prevention of ADRs.
5 CONCLUSION
This study revealed that the SNPs of XRCC1, ABCB1, CYP3A5 and GSTP1 were related to the hematotoxicity, particularly severe neutropenia and leukopenia, in patients undergoing platinum-based chemotherapy. In the current era of genetic diagnosis and treatment, SNPs play nonnegligible roles in predicting and preventing ADRs. As a part of this approach, these SNPs contribute to the development of individualised medicinal treatment. Therefore, additional studies are needed to identify the SNP-ADR relationship, which will facilitate the development of the personalised chemotherapy for cancer patients, thereby reducing the risk of adverse reactions and economic burden.
AUTHOR CONTRIBUTIONS
Shilong Zhong and Shuyao Zhang designed the study, led the study and provided resources. Shuang Chen designed the study, performed experiment and analysed data, and was a major contributor in writing the manuscript. Xiao Xiao revised the manuscript and provided resources. Jianliang Chen participated in patient recruitment and curated the data. Yun Chen performed the experiment and revised the manuscript. Zixian Wang performed data analysis. Guodong Qiu made the investigation. All authors reviewed and approved the final manuscript.
ACKNOWLEDGMENTS
There is no Acknowledgements. This study was supported by the Medical Scientific Research Foundation of Guangdong Province of China (A2022446), Science and Technology Development Projects of Guangzhou (202201011424), the Red Cross Foundation of China, New exploration and scientific research project of sepsis target therapy of Special fund for medical empowerment (HSZH202200909), the Guangdong Health Charity Foundation, 2022 Charity and Public Welfare Medical Clinical Research Project (JZ2022008), Research-oriented Hospital Program of Guangzhou (RHPG05), the Primary Health Care Foundation of China-Clinical Application Research and Medical Training Fund Project Key Project (2021120002) and Talent Project established by Chinese Pharmaceutical Association Hospital Pharmacy department (CPA-Z05-ZC-2021-003).
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
ETHICS STATEMENT
This study was approved by the Ethics Committee of the Cancer Hospital Affiliated to Shantou University School of Medicine (Approval number: 201809) and the Ethics Committee of Guangzhou Red Cross Hospital (Approval number: 2020-109-02), and conducted according to the Declaration of Helsinki.
INFORMED CONSENT
Informed consent was obtained from all the individual participants included in the study.
Open Research
DATA AVAILABILITY STATEMENT
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.




