Targeting KRAS in Cancer Therapy: Beyond Inhibitors
Funding: This work was supported by the Guangdong Basic and Applied Basic Research Foundation (2022A1515010520), the Guangzhou Municipal Science and Technology Project (2023A03J0758), and the Specific Research Fund for TCM Science and Technology of Guangdong Provincial Hospital of Chinese Medicine (YN2022DB01).
A recent study by Popow et al. published in Science described a heterobifunctional small-molecule pan-KRAS degrader ACBI3 [1], while two studies from the SUN group, published in Developmental Cell and MedComm, collectively elucidated the protein homeostasis mechanism of DIRAS2 and proposed strategies to indirectly target KRASG12D through physiological negative regulators [2, 3]. These studies provide novel perspectives on targeting KRAS in cancer therapy beyond inhibitors.
KRAS mutations are prevalent in human cancers. These mutations prevent GTP hydrolysis, leading to sustained RAS activation, downstream signal transduction, and tumorigenesis. For decades, KRAS was considered “undruggable” due to its smooth structure, picomolar affinity for GTP, and the lack of binding pockets on the GTP-bound form. This changed when the Food and Drug Administration approved sotorasib and adagrasib for the treatment of KRASG12C non-small cell lung cancer (NSCLC). In recent years, inhibitors targeting G12C (Olomorasib, Opnurasib, Glecarisib, IBI351, Garsorasib, RMC-6291, FMC-376, and D3S-001), G12D (MRTX1133, RMC-9805), and pan-RAS (RMC-6239, BI-3706674) have advanced into clinical trials [4]. Notably, IBI351 and Garsorasib were approved by the National Medical Products Administration in 2024 for the treatment of KRASG12C mutant NSCLC. However, no breakthroughs have yet been achieved for KRAS mutations other than G12C.
Proteolysis-targeting chimeras (PROTACs), which recruit E3 ubiquitin ligases to target proteins for degradation via the ubiquitin-proteasome pathway, have revolutionized drug development for challenging targets like KRAS. Popow et al. developed a PROTAC linking KRAS to the E3 ubiquitin ligase CRL2VHL using a high-affinity KRAS switch II pocket ligand previously disclosed in Nature. The molecule was optimized over three rounds. First, they replaced the oxygen in the linker with a methylene group to improve both cellular permeability and VHL affinity. Next, they substituted the amide with isoxazole to enhance the degradation efficiency of KRAS variants. Finally, they swapped isoxazole for triazole and added a hydroxymethyl group at the benzylic position of the VHL binder to raise in vivo active concentrations. The final molecule, ACBI3, degrades 13 of the 17 most common oncogenic KRAS variants, exhibits high-affinity VHL engagement (binary complex half-life >2000 s), and improves ternary complex stability (Kd = 6 nM). This drives potent KRAS degradation (half-maximal degradation concentration = 3.9 nM in GP2d cells) and selective antiproliferative activity in KRAS mutant cells (half-maximal inhibitory concentration = 478 nM), leading to significant tumor regressions in vivo. However, ACBI3 targets both mutant and wild-type KRAS, which may pose potential risks, even though Popow et al. demonstrated that compound 4, the precursor of ACBI3, does not affect the cell cycle or proliferation of wild-type KRAS cell lines, and ACBI3 administration in vivo does not alter mouse body weight.
In addition to PROTAC, scientists have identified new targets by elucidating the regulatory mechanisms of the KRAS signaling pathway. Recently, the SUN group uncovered the protein homeostasis mechanism of DIRAS2, a poorly studied member of the DIRAS family. DIRAS2 exhibits low GTPase activity and primarily exists in a GTP-bound form, functioning as a negative regulator of RAS. Specifically, Chang et al. discovered that ubiquitin-conjugating enzyme E2 F (UBE2F), a NEDD8-conjugating enzyme (E2) that specifically activates Cullin-RING ligase 5 (CRL5) through a cascade reaction, is overexpressed in pancreatic ductal adenocarcinoma (PDAC) and associated with poor prognosis. Knockdown of UBE2F inhibits the growth and survival of KRAS mutant pancreatic cancer cells. Using a pancreas-specific UBE2F conditional knockout (CKO) mouse model and 3D culture, they demonstrated that UBE2F deletion suppresses pancreatic tumorigenesis. After confirming that UBE2F positively regulates mitogen-activated protein kinase (MAPK) signaling, they identified DIRAS2 as a target of UBE2F, showing that UBE2F depletion leads to CRL5 inactivation and DIRAS2 accumulation. Through in vivo and in vitro ubiquitination assays, they revealed that DIRAS2 is a substrate of CRL5 and is subjected to K11-linked polyubiquitination. Using siRNA-based screening of all known CRL5 receptor subunit proteins, they identified ankyrin repeat and SOCS box protein 11 (ASB11) as the substrate receptor for CRL5 that destabilizes DIRAS2. Furthermore, they demonstrated that DIRAS2 disrupts RAS-RAF binding in a KRAS mutant-dependent manner. The accumulation of DIRAS2 induced by UBE2F deletion suppresses pancreatic lesion development in UBE2F and DIRAS2 CKO mouse models. In contrast to UBE2F, higher DIRAS2 levels correlate with better survival in PDAC patients. Shortly after Chang et al. identified CRL5ASB11 as the E3 ligase for DIRAS2, Chen et al. identified ubiquitin-specific protease 10 (USP10) as its deubiquitylase through affinity purification and mass spectrometry analysis. Using in vivo ubiquitination assays, they demonstrated that USP10 cooperates with CRL5ASB11 to regulate DIRAS2 stability and pancreatic cancer cell growth. These two studies collectively elucidate the mechanism of DIRAS2 ubiquitination and its role in the tumorigenesis of KRASG12D pancreatic cancer, offering several promising therapeutic targets for KRASG12D PDAC, including NEDD8 activating enzyme, UBE2F, sensitive to apoptosis gene (SAG)/RING Box protein 2 (RBX2) and ASB11 (Figure 1). However, several questions remain unresolved, such as why DIRAS2 selectively targets mutant KRAS over wild-type RAS and whether other DUBs influence DIRAS2 activity.
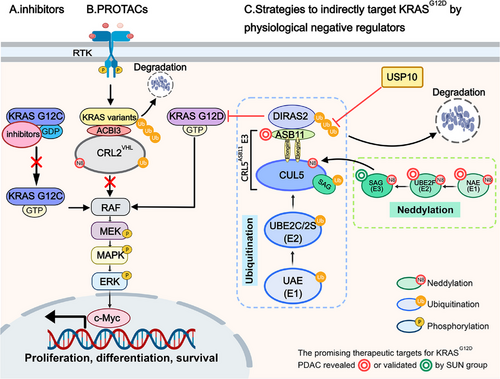
In summary, target-driven PROTACs enable direct KRAS degradation through rational engineering, offering a unique approach to target “undruggable” proteins with high efficiency and potential to overcome drug resistance. However, they face challenges such as high hydrophobicity, poor cellular permeability, and limited tumor-targeting ability, often requiring iterative optimization cycles, resulting in significant time, financial, and labor costs. The recently reported quantum-classical generative model demonstrates a notable advantage in experimentally validated hits over classical models for KRAS inhibitor discovery [5]. This improved performance likely arises from quantum effects, such as superposition and entanglement. A broader application of this approach could significantly accelerate PROTAC optimization.
Unlike the well-established inhibitors or the rapidly advancing PROTACs targeting KRAS, studies from the SUN group present a novel insight: indirectly and selectively targeting KRAS mutants by physiological negative regulators, such as DIRAS2. This approach involves more uncertainties, and its clinical translation may be prolonged due to complexity. However, it is this very uncertainty that drives a deeper understanding of KRAS regulatory mechanisms, which could even lead to breakthroughs similar to the identification of the switch II pocket, potentially overcoming the long-standing challenges in targeting KRAS.
Author Contributions
C.C. wrote the manuscript and prepared the figure. Z.P. provided valuable discussion. L.Z. revised and finalized the manuscript. All authors have read and approved the final manuscript.
Acknowledgments
This work was supported by the Guangdong Basic and Applied Basic Research Foundation (2022A1515010520), the Guangzhou Municipal Science and Technology Project (2023A03J0758), and the Specific Research Fund for TCM Science and Technology of Guangdong Provincial Hospital of Chinese Medicine (YN2022DB01).
Ethics Statement
The authors have nothing to report.
Conflicts of Interest
The authors declare no conflict of interest.
Open Research
Data Availability Statement
Not applicable.




