Whole-genome comparison between the type strain of Halobacterium salinarum (DSM 3754T) and the laboratory strains R1 and NRC-1
Graphical Abstract
The genome of the Halobacterium salinarum type strain (91-R6, DSM 3754) was compared at the DNA and protein levels to the genomes of two well-studied laboratory strains, NRC-1 and R1. The chromosomes and portions of the plasmids were very closely related. However, distinct homologs for proteins involved in motility and N-glycosylation were encountered.
Abstract
Halobacterium salinarum is an extremely halophilic archaeon that is widely distributed in hypersaline environments and was originally isolated as a spoilage organism of salted fish and hides. The type strain 91-R6 (DSM 3754T) has seldom been studied and its genome sequence has only recently been determined by our group. The exact relationship between the type strain and two widely used model strains, NRC-1 and R1, has not been described before. The genome of Hbt. salinarum strain 91-R6 consists of a chromosome (2.17 Mb) and two large plasmids (148 and 102 kb, with 39,230 bp being duplicated). Cytosine residues are methylated (m4C) within CTAG motifs. The genomes of type and laboratory strains are closely related, their chromosomes sharing average nucleotide identity (ANIb) values of 98% and in silico DNA–DNA hybridization (DDH) values of 95%. The chromosomes are completely colinear, do not show genome rearrangement, and matching segments show <1% sequence difference. Among the strain-specific sequences are three large chromosomal replacement regions (>10 kb). The well-studied AT-rich island (61 kb) of the laboratory strains is replaced by a distinct AT-rich sequence (47 kb) in 91-R6. Another large replacement (91-R6: 78 kb, R1: 44 kb) codes for distinct homologs of proteins involved in motility and N-glycosylation. Most (107 kb) of plasmid pHSAL1 (91-R6) is very closely related to part of plasmid pHS3 (R1) and codes for essential genes (e.g. arginine-tRNA ligase and the pyrimidine biosynthesis enzyme aspartate carbamoyltransferase). Part of pHS3 (42.5 kb total) is closely related to the largest strain-specific sequence (164 kb) in the type strain chromosome. Genome sequencing unraveled the close relationship between the Hbt. salinarum type strain and two well-studied laboratory strains at the DNA and protein levels. Although an independent isolate, the type strain shows a remarkably low evolutionary difference to the laboratory strains.
1 INTRODUCTION
Halobacterium salinarum is a rod-shaped, motile, extremely halophilic archaeon (Class Halobacteria) which grows best at NaCl concentrations in the range of 3.5–4.5 M (Grant, Kamekura, McGenity, & Ventosa, 2001). Members of this species are aerobic heterotrophs found in hypersaline environments worldwide, such as salt lakes and solar salterns, and often contaminate commercial preparations of raw (unprocessed) solar salt (Henriet, Fourmentin, Delince, & Mahillon, 2014). It has been extensively studied as a model archaeal extremophile, resulting in numerous discoveries and insights into archaeal biology and the adaptations required to live at saturating salt concentrations (see reviews by Beer, Wurtmann, Pinel, & Baliga, 2014; Soppa, 2006) and the references within). Examples include prokaryotic glycoproteins (Mescher & Strominger, 1976), archaeal isoprenoid lipids and membranes (Kellermann, Yoshinaga, Valentine, Wormer, & Valentine, 2016), rhodopsins (Grote & O'Malley, 2011), resistance to UV-induced DNA damage (Jones & Baxter, 2017), gene transcription and regulation (Yoon et al., 2011), motility via archaella (Kinosita, Uchida, Nakane, & Nishizaka, 2016), biofilm formation (Fröls, Dyall-Smith, & Pfeifer, 2012), halovirus biology (Stolt & Zillig, 1993), and even astrobiology (Leuko, Domingos, Parpart, Reitz, & Rettberg, 2015). Unusual features of this species are the high level of genetic variation, due mainly to the presence and activity of numerous ISH elements (Brugger et al., 2002), and the high GC content of the main chromosome (~68%) compared to their plasmids (57%–60% G + C) (Grant et al., 2001; Ng et al., 2000; Pfeiffer, Schuster, et al., 2008).
Halobacterium salinarum was first isolated in 1922 from cured cod by Harrison and Kennedy, who named it Pseudomonas salinaria (Harrison & Kennedy, 1922). The source of this organism was found to be salt. The original type strain of Hbt. salinarum was lost and, as described by Grant (Grant et al., 2001), a neotype was assigned as Hbt. salinarum isolate 91-R6 (Lochhead, 1934), which is maintained in several culture collections (NRC 34002 = ATCC 33171 = DSM 3754 = JCM 8978 = NCMB 764 = CIP 104033 = NBRC 102687) and which we refer to as strain 91-R6 hereafter. The neotype was isolated in Canada from the red discoloration found on a salted cowhide (Lochhead, 1934). Similar isolates from this and other sources were reported over the years and variously named Hbt. salinarum, Hbt. cutirubrum, or Hbt. halobium but were later found to be so closely related that those named Hbt. cutirubrum and Hbt. halobium were transferred to the salinarum species (Ventosa & Oren, 1996). Detailed taxonomic descriptions of the Order Halobacteriales are given in (Grant et al., 2001; Gupta, Naushad, & Baker, 2015; Oren, 2006, 2014).
The previously sequenced Hbt. salinarum strains R1 and NRC-1 are most likely derived from the isolate DSM 670 (Gruber et al., 2004), which is supported by their closely similar genome sequences (Pfeiffer, Schuster, et al., 2008). Both have a 2 Mb main chromosome. Their plasmids share 350 kb of near-identical unique sequence despite major differences in overall plasmid arrangement: strain NRC-1 carries two (191 and 365 kb) and R1 four (41, 148, 195 and 284 kb) plasmids (Ng et al., 2000; Pfeiffer, Schuster, et al., 2008). Both sets of plasmids are correctly assembled as evidenced by the available experimental data for strain R1 (Pfeiffer, Schuster, et al., 2008) and for strain NRC-1 (Bobovnikova, Ng, Dassarma, & Hackett, 1994; Kennedy, 2005; Ng, Arora, & Dassarma, 1993; Ng et al., 2008, 1998, 2000; Ng & DasSarma, 1991; Ng, Kothakota, & Dassarma, 1991). While strain DSM 670 is thought to derive from NRC 34020, the original source and isolation details appear to be lost. Taken together, and from the information available, it could be anticipated that the type strain 91-R6 (DSM 3754T = NRC 34002) is an independent isolate compared to strains R1 and NRC-1.
In 2012, Oren pointed out that even though Hbt. salinarum DSM 3754T is taxonomically important as the “type species of the type genus of the family and the order,” its genome had not been sequenced (Oren, 2012). An incomplete sequencing project is listed in the JGI GOLD database (Gp0108295), but access is restricted.
We have determined the complete genome sequence of the type strain of Hbt. salinarum (strain 91-R6; DSM 3754T) using long-read PacBio sequencing (Pfeiffer, Marchfelder, Habermann, & Dyall-Smith, 2019). Here we describe its characteristics in more detail and then focus on its relationship to the widely studied laboratory strains R1 and NRC-1 at the DNA and protein levels.
2 MATERIALS AND METHODS
2.1 Cell cultivation and genome sequencing
Cells of the type strain of Hbt. salinarum (strain 91-R6; DSM 3754T) were obtained from the DSMZ and were inoculated into liquid complex medium omitting any colony purification. The medium contained 250 g/L (w/v) NaCl, 20 g/L MgSO4·7H2O (w/v), 3 g/L sodium citrate (w/v), 2 g/L KCl (w/v), and 10 g/L peptone (w/v) (Oxoid LP0034) and was adjusted to pH 7. Cells were grown aerobically at 37°C under shaking (105 rpm, Innova 43) to an OD600 of 0.3. A sample was withdrawn for PCR validation before the cells were pelleted by centrifugation for 8 min at 5,100 g at room temperature. Cell pellets were frozen and stored at −80°C.
The authenticity of the collected cells was validated by sequencing of PCR products for the 16S rRNA and rpoB genes using primers listed in Table 1. Chromosomal DNA was isolated using the spooling method as described for Haloferax volcanii in the Halohandbook (Dyall-Smith, 2009). PCR fragments were generated and analyzed by Sanger sequencing.
| Primer | Sequence |
|---|---|
| 16SHabc#1 | 5′-CTGCGGTTTAATTGGACTCAACGCC-3′ |
| 16SHabc#2 | 5′-GATTCCCCTACGGCTACCTTGTTAC-3′ |
| BrpoB2Vorn1 | 5′-CCTCCGGGCAGGGCAAGAACTACCAG-3′ |
| BrpoB2Hinten1 | 5′-GCGAAGTTCTTCACCAGCCCACAGTT-3′ |
After validation, the cell pellet was sent to the Max-Planck Genome Center Cologne (https://www.mpgc.mpipz.mpg.de) for DNA extraction, library preparation, and sequencing as reported previously (Pfeiffer et al., 2019). The sequence was determined with a PacBio RSII instrument (Rhoads & Au, 2015). The kits from PacBio were used according to the manufacturer's instructions (DNA template preparation kit; DNA/polymerase binding kit; DNA sequencing kit; MagBead kit; SMRT cell 8pac).
2.2 Genome assembly
As reported previously (Pfeiffer et al., 2019), an initial and automated genome assembly was performed at the Max-Planck Genome Center Cologne, using the SMRTanalysis pipeline (PacificBiosciences) which runs HGAP (DAGCON-based hierarchical genome assembly process, RS_HGAP_assembly.2 version 2.3.0) with the following three steps: preassembly, de novo assembly with the Celera assembler and final polishing with Quiver. The data originated from five SMRT cells. We obtained 253,044 reads with an average length of 5,400 bp (1 Gbp total). Despite extremely high coverage (>400-fold), the assembly resulted in 43 distinct contigs. A supervised genome assembly was then applied using CANU v1.7 (Koren et al., 2017) for assembly and Geneious v10.2 (Kearse et al., 2012) (www.geneious.com) for integration and editing of contigs. This allowed the correct handling of genomic polymorphisms, and enabled closure of all replicons (one chromosome and two large plasmids), resulting in a representative genome.
2.3 Analysis of genome heterogeneity
Various polymorphisms were encountered in the original PacBio reads, which were found to be associated with mobile genetic elements (MGE) and were responsible for the failure of the automated genome assembly. To analyze these, 150 bp of unique sequence was selected on each side of the polymorphic MGE, concatenated, and then compared (BLASTn) against the entire set of PacBio reads. Blast hits better than E = 10–20 were analyzed by visual inspection. PacBio reads were categorized according to the type of connectivity they exhibited, as (a) contiguous, (b) split by the MGE but otherwise consistent with the assembly, or (c) indicative of a rearrangement compared to the representative genome.
2.4 Genome comparison strategy
We recently described the comparison of two closely related strains of Photorhabdus laumondii (Zamora-Lagos et al., 2018) and adopted the same analysis strategy for Halobacterium. Briefly, matching segments (matchSEGs) were identified by an initial pairwise MAFFT (Katoh & Standley, 2013) alignment in chunks of 400 kb. These were subsequently fine-tuned in an iterative approach. Script-based checking ensured that matchSEGs did not contain indels larger than 100 bp. All regions with >4% sequence difference in a 1,000 bp window were manually checked to determine whether they represented contiguous matchSEGs with an elevated difference ratio, or were composed of distinct matchSEGs with an intervening strain-specific sequence.
For matchSEGs, sequence similarity statistics were computed from the MAFFT alignment by a custom script. Each position was classified to be a “match” (m), a “mismatch” (mm), a “gap open” (go), or a “gap extension” (ge) position. Gap extension positions were excluded from subsequent computations. Sequence difference was calculated using the formula (mm + go)/(m + mm+go).
Adjacent matchSEGs are separated by a divergent segment (divSEG) in at least one of the strains. DivSEGs were classified into two categories, indel or replacement (see text for more details). After completion of the analysis, a custom script verified that each genome position is classified exactly once, either as part of a matchSEG or part of a divSEG. All MAFFT alignments were confirmed to represent the specified genomic region. All matchSEGs were checked to confirm that there were no base mismatches at their first and last alignment positions.
2.5 Further enhancement of the annotation of protein-coding genes in the Hbt. salinarum R1 genome
The annotation of the Hbt. salinarum R1 genome reflects an extensive Gold Standard Protein based manual curation (Pfeiffer & Oesterhelt, 2015) and is used as a reference for the strain 91-R6 annotation. This annotation is regularly and systematically kept up-to-date, based on principles published in 2015 (Pfeiffer & Oesterhelt, 2015). This also includes regular systematic correlation with a high-level database (SwissProt). Our procedures have been extended in the context of the current study to additionally include a detailed and systematic comparison to the KEGG database annotation (Kanehisa, Sato, Furumichi, Morishima, & Tanabe, 2019). The genes represented in KEGG for Hbt. salinarum R1, Hfx. volcanii, and Natronomonas pharaonis were downloaded. In KEGG, proteins are only annotated when they are assigned to a KO (Kegg Orthology). For these, protein names and EC numbers were compared between the two annotation systems. If the KEGG annotation was considered superior (e.g. (a) is consistent with a recent revision of the EC number assignment; (b) assigns a specific function, including published evidence), we updated our own annotation. If we considered our annotation superior, we sent feedback to KEGG. In our report of the manual curation strategy (Pfeiffer & Oesterhelt, 2015), we have pointed to the severe problems caused by overannotation (assignment of a specific protein function while there is only support for a general function assignment; see also Schnoes, Brown, Dodevski, and Babbitt (2009) for this subject). We consider some of the specific function assignments by KEGG as overannotations, which seems to be caused by relaxed conditions for some of the KEGG orthology assignments. Based on our annotation principles, we only assign a general function in such cases but are aware that KEGG applies the opposite annotation policy in such cases.
2.6 Annotation of protein-coding genes from the Hbt. salinarum 91-R6 genome
Gene prediction was initially performed using GeneMarkS-2 (Lomsadze, Gemayel, Tang, & Borodovsky, 2018). Proteins with sequence identity between strains 91-R6 and R1 were correlated by a custom PERL script. All noncorrelated sequences from strain 91-R6 were compared to the ORF set from strain R1 by BLASTp. The majority of proteins could be correlated by this method, and typically had 99% protein sequence identity. It has been shown previously that start codon assignments are highly unreliable for GC-rich genomes (Falb et al., 2006). All obvious start codon assignment discrepancies detected upon BLASTp result analysis were resolved by manual curation, applying published procedures (Pfeiffer & Oesterhelt, 2015). Disrupted genes, which became evident at this stage, were subjected to manual curation. In order to minimize missing gene calls, all intergenic regions (≥50 bp) in the strain 91-R6 genome were confirmed as noncoding by using BLASTx searches against (a) a protein set from 12 haloarchaeal genomes, including that from Hbt. salinarum strain R1 (Pfeiffer & Oesterhelt, 2015) and (b) NCBI:nr. All strain-specific proteins were analyzed by tBLASTn to ensure that they are not encoded in the partner genome. Missing genes, which were detected by this analysis, were postpredicted and thus resolved.
For correlated proteins, the annotation from the reference strain R1 was copied to the strain 91-R6 protein. All strain-specific proteins were annotated by comparison to (a) the set of carefully annotated haloarchaeal genomes (Pfeiffer & Oesterhelt, 2015), (b) the SwissProt section of UniProt, and (c) the TrEMBL section of UniProt and the associated InterPro domains.
2.7 Third party annotation of protein-coding genes from the Hbt. salinarum NRC-1 genome
The genomes of strains R1 and NRC-1 are exceedingly similar (Pfeiffer, Schuster, et al., 2008), and in genome regions with complete sequence identity their predicted protein-coding genes should be identical. Where necessary, the NRC-1 start codons were reassigned to match those from the extensively curated genes of strain R1. Also, protein names, genes, and EC numbers were updated for NRC-1 if required. In cases where corresponding genes had mutated but retained >99% sequence identity at the DNA level, the NRC-1 gene was annotated to best correlate with the R1 gene. NRC-1 has only 15 kb of unique sequence which is not represented in the R1 genome and these regions were annotated according to our established procedures (Pfeiffer & Oesterhelt, 2015).
2.8 Annotation of stable RNAs in all three strains
All stable RNA gene coordinates (rRNAs, tRNAs, RNase P RNA, 7S RNA) were brought into line with their annotation in RFAM (Kalvari et al., 2018). For Halobacterium, the stable RNA annotations from strain NRC-1 (taxid: 64091) are reported in RFAM and were kindly provided by RFAM staff (obtained Feb-2019).
First, the RNA annotations in strain R1 were curated. All RNA function assignments were found to be consistent with RFAM, while coordinates deviated for several RNAs. This was resolved by using BLASTn analyses with the RFAM-provided NRC-1 RNAs. For some tRNAs, which are not represented in the RFAM annotation of NRC-1, coordinates could be reliably delineated from homologous tRNAs.
For strains 91-R6 and NRC-1, stable RNAs were subsequently adjusted to those from strain R1, based on BLASTn analyses.
2.9 Transposon analysis
Transposons were identified by BLASTn and BLASTx comparison to an extensive in-house collection of haloarchaeal transposons and to the ISFinder database (Siguier, Perochon, Lestrade, Mahillon, & Chandler, 2006; Siguier, Varani, Perochon, & Chandler, 2012) by a previously described procedure (Pfeiffer et al., 2018). Identified transposons were added to the in-house database and were used for a subsequent iterative transposon analysis using BLAST. Newly identified transposons were submitted to and have been accepted by ISFinder. In addition to canonical transposons, we identified several MITEs (Miniature Inverted-Terminal-repeat Elements), which were submitted to and accepted by ISFinder for their recently introduced MITE subsection.
2.10 Additional bioinformatics tools
As general tools, MUMMER v4 (Delcher, Salzberg, & Phillippy, 2003) and the BLAST suite of programs v2.9 (Altschul et al., 1997; Johnson et al., 2008) were used for genome comparisons. The CRISPR finder web server (http://crispr.i2bc.paris-saclay.fr) was used to search for CRISPR elements (Grissa, Vergnaud, & Pourcel, 2008). Prophage searches were performed online using PHASTER (http://phaster.ca) and Profinder (http://aclame.ulb.ac.be/Tools/Prophinder). In silico DNA-DNA hybridization (DDH) values were calculated using the Genome-to-Genome Distance Calculator (GGDC) 2.1 server at http://ggdc.dsmz.de/ggdc.php. ANIb (average nucleotide identity, BLASTn) values were determined using the JSpecies server at http://jspecies.ribohost.com/jspeciesws. Circular genome maps were created using the CGView Server (http://stothard.afns.ualberta.ca/cgview_server). Genomic island (GI) prediction used Island Viewer 4 (http://www.pathogenomics.sfu.ca/islandviewer) described by Bertelli et al. (2017).
3 RESULTS
3.1 Genome sequencing and assembly for Hbt. salinarum strain 91-R6
3.1.1 Cell cultivation, genome sequencing strategy, and closing of the replicons
As Halobacterium is known to be a genetically unstable organism (DasSarma et al., 1988; Pfeifer & Blaseio, 1989; Pfeifer, Weidinger, & Goebel, 1981), we avoided microbial manipulations (colony purification) that would select a clonal population for sequencing. A freshly obtained sample of Hbt. salinarum strain 91-R6 (DSM 3754T) was directly inoculated into liquid growth medium and, after expansion to the required amount of cellular material and removal of a sample for validation, cells were collected by centrifugation, frozen, and stored at −80°C. After validation of the strain by PCR analysis of 16S rRNA and the rpoB gene (for primers see Table 1), the frozen cells were transferred to the sequencing center for DNA extraction, library preparation, genome sequencing, and automated genome assembly.
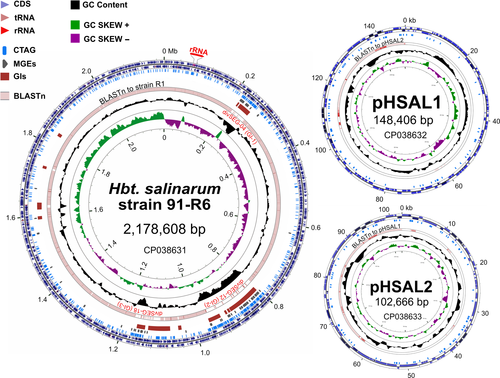
The genomes of previously sequenced Halobacterium strains had been very difficult to assemble because they carry numerous transposons and very long duplications in their plasmids. In the current study, PacBio long-read sequencing technology with very high sequence coverage (>400-fold) was chosen specifically to overcome these problems, but the automated assembly still failed to close the replicons, and 43 contigs were obtained. A supervised assembly process allowed closure, resulting in a representative genome with three circular replicons: a main chromosome (2,178,608 bp, 67.1% GC) and two large plasmids (pHSAL1, 148,406 bp, 60.6% GC; pHSAL2, 102,666 bp, 56.5% GC). The plasmids share a perfect duplication of 39,230 bp. The overall genomic arrangement of a highly GC-rich chromosome with less GC-rich plasmids that carry extensive duplications is similar to that found in other Halobacterium strains (Jaakkola et al., 2016; Lim et al., 2016; Ng et al., 2000; Pfeiffer, Schuster, et al., 2008) (see also Appendix 3).
The failure of the automated assembly process was due to a significant level of genomic population heterogeneity (see below, Section 3.3), which was associated with mobile genetic elements (MGEs). The representative genome includes all unique sequences that were obtained, but does not include those transposon copies which are found in only part of the population. A very close relationship between the chromosome of strain 91-R6 and those of the laboratory strains R1 and NRC-1 was immediately obvious, and is described in detail below (Section 3.2). Due to the extreme similarity between the chromosomes of strains R1 and NRC-1 (only 12 differences aside from MGE targeting and MGE-internal sequence differences), the NRC-1 chromosome is fully covered by analyzing the R1 chromosome.
The plasmids of strains R1 and NRC-1 vary in number and gene arrangement and thus both are included in the comparative analysis. However, all the unique sequences shared between the R1 and NRC-1 plasmids are near-identical.
3.1.2 Setting the point of ring opening for each replicon
After finalization of the genome assembly, a starting base was set for each of the three circular replicons. For the chromosome, we adopted the convention of choosing a position close to a canonical replication origin. However, we used a biologically relevant variation that we have used previously for Natronomonas moolapensis (Dyall-Smith et al., 2013) and Halobacterium hubeiense (Jaakkola et al., 2016). Most haloarchaeal genomes contain a canonical replication origin that is flanked on one side by a distinctive, highly conserved paralog of the Orc/Cdc6 family, and on the other side by a highly conserved but divergently transcribed three-gene cluster (oapABC; oap: origin-associated protein). The highly conserved, origin-associated Orc paralog can be considered the functional equivalent of the bacterial dnaA gene, which is typically the 1st gene on a bacterial chromosome. Equivalently, in many haloarchaea, the ring is opened upstream of that Orc paralog, with the Orc paralog assigned to the forward strand. However, this breaks the Orc/oapABC junction, the latter ending up as the last three genes of the chromosome. In the genome representation selected by us, the chromosome is opened on the other side of the oapABC cluster to avoid the disjunction between oapABC from the replication origin and the associated Orc gene. The Orc gene thus becomes the 4th gene of the chromosome, being encoded on the forward strand.
The plasmid rings were opened so that both plasmids terminate with the perfect 39,230 bp duplication.
An overview of the replicons of the analyzed strains, including summary data for the plasmids and the complete genome, is shown in Tables 2 and 3. Strain 91-R6 follows the same pattern already observed for the laboratory strains: a GC-rich chromosome of ~2 Mb accompanied by megaplasmids (or minichromosomes) of diminished GC content and with large-scale duplications. The three replicons are depicted in Figure 1. Further details on the chromosomes and plasmids from the three analyzed strains are provided in Appendix 3.
| Replicon | 91-R6 | R1 | NRC-1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | pHSAL1 | pHSAL2 | Chr | pHS1 | pHS2 | pHS3 | pHS4 | Chr | pNRC100 | pNRC200 | |
| Length (bp) | 2,178,608 | 148,406 | 102,666 | 2,000,962 | 147,625 |
194,963 |
284,332 |
40,894 |
2,014,239 | 191,346 | 365,425 |
| GC (%) | 67.1 | 60.6 | 56.5 | 68.0 | 57.4 | 58.6 | 59.8 | 57.9 | 67.9 | 57.9 | 59.2 |
| #Proteins | 2,346 | 170 | 108 | 2,151 | 168 | 220 | 291 | 38 | 2,174 | 223 | 420 |
| #Pseudo | 106 | 37 | 30 | 43 | 19 | 27 | 65 | 1 | 59 | 39 | 73 |
| #RNAs | 53 | – | – | 52 | – | – | – | – | 52 | – | – |
Note
- Core statistical data are given for the replicons of strain 91-R6 and those of strains R1 and NRC-1. Replicon Chr refers to the main chromosome, and plasmids are represented by their names. The number of RNAs (#RNAs; rRNAs + tRNAs + ncRNAs) is given. For proteins, the total number (#proteins) and the number of disrupted proteins (pseudogenes, “#pseudo”) are given. For R1, the protein-related data differ slightly from the original publication (Pfeiffer, Schuster, et al., 2008) due to subsequent annotation updates. Protein-coding genes which have been targeted by a MGE are represented (and thus counted) as a single multiregion ORF in strain R1 and as multiple single-region ORFs in strains 91-R6 and NRC-1 (for details see Appendix 4).
| 91-R6 | R1 | NRC-1 | ||||
|---|---|---|---|---|---|---|
| Plasmids | Genome | Plasmids | Genome | Plasmids | Genome | |
| Length (bp) | 251,072 | 2,429,680 | 667,814 | 2,668,776 | 556,771 | 2,571,010 |
| GC (%) | 58.9 | – | 58.8 | – | 58.8 | – |
| #Proteins | 278 | 2,624 | 717 | 2,868 | 643 | 2,817 |
| #Pseudo | 67 | 173 | 112 | 155 | 112 | 171 |
| #RNAs | – | 53 | – | 52 | – | 52 |
Note
- The data presented for each replicon (see Table 2) are summarized here as aggregate values for all plasmids of each strain and for the complete genome (chromosome plus all plasmids). Duplicated protein-coding genes on plasmid region duplications are counted several times.
3.1.3 Genome features
DNA methylation
Using the PacBio reads and the assembled genome sequence, base modifications were analyzed using the SMRT® Analysis software (Basemods tool) (Chin et al., 2013). All replicons contained methylated C residues (m4C) at position 1 of the tetranucleotide sequence CTAG, on both strands. Methylation was estimated to be present at >90% of sites. The CTAG motif was significantly under-represented in all three replicons; a feature that is commonly found in many haloarchaeal genomes (Fullmer, Ouellette, Louyakis, Papke, & Gogarten, 2019). For example, there were only 1,430 sites on the chromosome (odds ratio = 0.37). Methylation is probably carried out by the Zim CTAG modification methylase (HBSAL_08190), a homolog of the methylase (HVO_0794) described for Hfx. volcanii (Hartman et al., 2010; Ouellette, Gogarten, Lajoie, Makkay, & Papke, 2018). The distribution of CTAG motifs around the chromosome and plasmids of strain 91-R6 is indicated in Figure 1 (see below).
Overall structure of the replicons
For the chromosome, a cumulative GC-skew plot (Figure 1, inner-most ring) shows an overall trend of increasing GC-skew while moving clockwise from the top, around the circle, and back to the top, with a strong inflection near the canonical replication origin (point of ring opening). This general pattern is similar to many bacterial genomes, where the major inflection point indicates the position of the replication origin (Lobry & Louarn, 2003). Variations in GC content (7th level, black plot) often coincide with disturbances of the GC-skew, as is seen across the single rRNA operon found close to and pointing away from the ori. Other, more extended regions of lower GC show higher densities of both MGEs (4th level, gray arrows) and CTAG motifs (3rd level, blue lines). A BLASTn comparison to strain R1 (6th level, pink) highlights the close similarity between the two strains, with only three large interruptions (labeled divSEGs 04, 12 and 18). Predicted genomic islands (GIs; 5th level, brown) are correlated with these divSEGs and represent likely regions of horizontally acquired DNA, and show the typical features of high levels of MGEs and lower than average GC. They also have a higher density of CTAG motifs. In summary, the chromosome appears to have an underlying organization, as evidenced by the cumulative GC-skew, interspersed by large genomic islands (HGT) and smaller indels.
Both plasmids (Figure 1, right side) have a reduced GC content compared to the chromosome; 6.5% less for pHSAL1 and 10.6% for pHSAL2 (Tables 2 and 3). The BLASTn rings of each map (4th level, pink) display the sequence similarity to the other plasmid, clearly revealing the 39.2 kb of sequence that they share in common. The unique region of plasmid pHSAL1 (107 kb) is near-identical to part of R1 plasmid pHS3 (see Figure A1 in Appendix 2).
Ribosomal RNA and tRNA genes
Strain 91-R6 has a single rRNA operon and 48 tRNA genes, all carried on the main chromosome. The rRNA operon has the typical bacterial gene order (Hui & Dennis, 1985): 16S–tRNAAla(UGC)–23S–5S–tRNACys(GCA), an arrangement noted previously in strains R1 and NRC-1 (Ng et al., 2000; Pfeiffer, Schuster, et al., 2008). The 16S and 5S rRNA sequences are identical to those of the R1 strain, while the 23S rRNA sequence differs by a single base change (nt 2,890, C/T). There are tRNAs for all 20 amino acids. Three tRNA genes contain predicted introns: tRNAIle(CAU), tRNATrp(CCA), and tRNAMet(CAU). The only tRNA difference between strains 91-R6 and R1 is that strain 91-R6 carries an extra (although partial) copy of tRNAGly(GCC) at nt 1,621,908–1,621,851, adjacent to a 7.5-kb indel (divSEG30, see below, Section 3.2).
3.1.4 Key physiological features of protein-coding genes
The annotation of protein-coding genes from all three strains has been extensively curated, and these genes have been correlated in great detail between strains (see Section 2 and Appendix 4, Appendix 5, Appendix 6). Here, we present physiological features of protein-coding genes which are prominently associated with Halobacterium (e.g. bacteriorhodopsin and motility). Due to extensive chromosomal sequence similarity, a majority of physiological features is common among the analyzed strains of Hbt. salinarum. We highlight those where we encountered differences and those which are otherwise relevant for archaeal biology.
Virus defence systems and prophage genes
No CRISPR regions or cas genes were detected in strain 91-R6. The R1 and NRC-1 strains also lack CRISPR-Cas genes (Ng et al., 2000; Pfeiffer, Schuster, et al., 2008). A search for other species of this genus that have sequenced genomes found complete CRISPR-Cas regions in two (Hbt. hubeiense and Halobacterium sp. DL1), a partial (and nonfunctional) system in one (Halobacterium jilantaiense), and none in Halobacterium noricense CBA1132. Recently, a distinct virus defense system has been identified in bacteria, the BREX system (Goldfarb et al., 2015), which is also present in many haloarchaea, including strain R1, where it is located on plasmid pHS3. Goldfarb et al. classified the haloarchaeal BREX system as “type 5.” Among the variations specific for this type, they identified a helicase domain gene, denoted as brxHII. While they were able to identify a helicase BrxHII in Haloarcula hispanica (HAH_4399), they did not identify this gene in Hbt. salinarum strain R1 (Goldfarb et al., 2015). The reason is that the gene (OE_5343R) is disrupted by transposon targeting and thus is not included in the protein sequence databases. BrxHII disruption may render the BREX system of Halobacterium nonfunctional, and this may be the reason why strain R1 (and its derivative S9) is susceptible to attack by viruses like phiH1 or ChaoS9 (Dyall-Smith et al., 2019; Dyall-Smith, Pfeifer, Witte, Oesterhelt, & Pfeiffer, 2018). This region of pHS3 is missing in strain NRC-1, which thus is devoid of a BREX system.
In strain 91-R6, distant homologs of the strain R1 BREX system proteins were identified, encoded by a cluster of closely spaced genes (brxABC and pglXZ; HBSAL_05050 to HBSAL_05080) on a strain-specific sequence of the chromosome (divSEG12, see below, Section 3.2). In strain 91-R6, no homolog to OE_5343R could be identified and no other helicase domain protein is encoded in the genomic vicinity to the BREX system. The pglX gene (DNA methyltransferase) is disrupted, and methylation analysis of the SMRT data did not indicate any motifs with methylation of A residues. At present, it is unclear whether the BREX system in this strain is functional.
Prophage prediction tools did not identify any integrated proviruses, but several strain-specific regions have characteristics which are typical for integrative elements (strain-specific regions with integrase genes in close vicinity to tRNA genes or having targeted a protein-coding gene and being bounded by a direct repeat) (see below, divSEG14, the divSEG15/16/17 trio, divSEG30, and divSEG31).
Opsin genes
Strain 91-R6 carries one bacteriorhodopsin (bop) gene, one halorhodopsin gene (hop), and two sensory rhodopsins (sopI, sopII). All are carried on the main chromosome along with their associated and regulatory genes (e.g. bat, bap, blp), and all are present in genomic regions strongly related to strain R1. The bop gene has a short insertion and may not be functional (see later, Section 3.2, divSEG27).
Motility genes
Archaellins (flagellins), the structural genes of the Halobacterium archaellum (flagellum), are encoded by a multigene family, and while the archaellin (flagellin) genes arlB1-B3 (previously flgB1-B3) are encoded in the type and both laboratory strains in immediate genomic vicinity to the motility (Arl, previously Fla) and chemotaxis (Che) clusters, the arlA1A2 (flgA1A2) gene pair of strain R1 is not associated with other motility or chemotaxis genes. Instead, this gene pair is encoded on a strain-specific sequence, as is a single arlA gene in strain 91-R6. Both arlA loci occur on divSEG18 (see below, Section 3.2). The protein sequence of ArlA is distinctly different from the homologs of other sequenced species, and by BLASTp was most similar (89% protein sequence identity) to ArlA (FlaA) of Hbt. jilantaiense (accession SEV92461.1).
N-glycosylation
In addition to the S-layer glycoprotein, there are many other haloarchaeal proteins that are known to be N-glycosylated, such as archaellins and some pilins (Jarrell et al., 2014). The pathway of N-glycosylation in Hbt. salinarum has also been studied (Kandiba & Eichler, 2015). Several enzymes of the N-glycosylation pathway (aglF, aglG, aglJ, aglM, aglR) are encoded as distant homologs on strain-specific regions (on divSEG18, see above “Motility genes” and below, Section 3.2). Strain 91-R6 lacks a close homolog of aglE. Additional glycosyltransferases are encoded in both strain-specific regions. The last bases of divSEG18 code for the N-terminal 18 codons of aglB (44% protein sequence identity), while the remainder of the protein is encoded on the subsequent matchSEG (98% protein sequence identity).
Biofilm formation
Strain 91-R6 is known to display a strong ability to form biofilms (Fröls et al., 2012; Losensky et al., 2017; Losensky, Vidakovic, Klingl, Pfeifer, & Frols, 2015). By comparison, strain R1 is nearly as proficient while strain NRC-1 shows negligible ability under the laboratory conditions tested. The close similarity of the genome sequences of these strains and their wide difference in biofilm phenotype attracted our attention, providing a basis for speculating on the genetic basis of biofilm formation in this species.
In Hfx. volcanii, PilA pilins are required for surface adhesion (Esquivel, Xu, & Pohlschroder, 2013). Several pilins of Haloferax are N-glycosylated, and interference with glycosylation has been shown to modify pilus assembly and function (Esquivel, Schulze, Xu, Hippler, & Pohlschroder, 2016). The six characterized PilA proteins of Haloferax share an identical 30 amino acid H-domain of their type III signal sequence. This represents a specific subtype of the more general Pilin_N (previously DUF1628) domain (PFAM:PF07790).
Strain 91-R6 has four proteins with an assigned DUF1628 domain, each with an ortholog in strains R1 and NRC-1. Strains R1 and NRC-1 have one additional, plasmid-encoded paralog. Curiously, only one DUF1628 domain protein (HBSAL_01455) has a type III signal sequence H-domain that is highly similar to Haloferax PilA. There are 22 strictly conserved residues, followed by relaxed similarity (three point mutations in eight residues). In the nonadhesive strain NRC-1, the ortholog is disrupted by transposon targeting (VNG_0110d + VNG_0112a), while the corresponding genes are intact in the adhesive strains R1 (OE_1186A1F) and 91-R6 (HBSAL_01455). The proteins from strain R1 and 91-R6 show 93% protein sequence identity, are identical in length (122 aa), and have four potential N-glycosylation sites. In Haloferax, the pilB3C3 gene pair has been identified as the PilA pilus assembly machinery (Esquivel & Pohlschroder, 2014). This assembly machinery is not clustered with its target genes, which in turn are not clustered with any assembly machinery. Most other pilBC assembly genes are in operons which also code for proteins with a type III signal sequence. For Halobacterium strain R1, it was shown that cells displayed a ten-fold reduction in glass adherence when the pilB1 gene was deleted (Losensky et al., 2015). Halobacterium pilB1 (OE_2215R, HBSAL_04190) is the ortholog of Haloferax pilB3 (HVO_1034) (same for Hbt. pilC1, OE_2212R, HBSAL_04185, vs. Hfx. pilC3, HVO_1033). From these analyses, we conclude that Halobacterium pilB1C1 is the assembly machinery for a nonclustered PilA pilin and that this PilA pilin mediates cell adhesion and the biofilm phenotype.
The enhanced biofilm formation properties of strain 91-R6 compared to R1 may be mediated either by protein sequence differences or by alterations in their N-glycosylation pathways (see above, N-glycosylation).
Amino acid biosynthesis genes
Halobacterium salinarum strain R1 (and NRC-1) is reported to be auxotrophic for several amino acids, including leucine and isoleucine (Falb et al., 2008; Gonzalez et al., 2009). However, strain 91-R6 codes for several genes of leucine and isoleucine/valine biosynthesis, specifically, leuABCD and ilvBCDN. The four genes ilvBCDN (within divSEG18, see below, Section 3.2) code for three enzymes with relaxed substrate specificity that catalyze equivalent reactions within the biosynthetic pathways of both isoleucine and valine. Consistent with bioinformatic reconstruction, strain 91-R6 grows well in the absence of leucine, isoleucine, and valine (Figure A3 in Appendix 2). While strain R1 did not grow in the absence of leucine, we observed growth in the absence of isoleucine and valine. This discrepancy between bioinformatic reconstruction and experimental results is yet unresolved. Besides the differences in isoleucine/valine and leucine biosynthesis genes, we did not detect any other differences in amino acid metabolism.
3.2 Detailed comparison of the type strain 91-R6 genome to that of strains R1 and NRC-1
3.2.1 Comparison of the chromosomes of strains 91-R6 and R1
Overall similarity between the chromosomes from the three strains of Hbt. salinarum and other species from the genus Halobacterium
The similarity between the chromosome of the type strain and the two laboratory strains was examined by in silico DNA–DNA hybridization (DDH) and average nucleotide identity (ANI), and the results are summarized in Tables A1 and A2 (Appendix 1). The type strain showed DDH values of 95% and ANI values of 98% to the laboratory strains, well above the accepted thresholds for membership of the same species (70% DDH; 95%–96% ANI) (Chun et al., 2018; Oren, Ventosa, & Grant, 1997). The ANI values also indicated a high level of sequence conservation between the strains. When compared to other recognized species of the genus Halobacterium, the type strain exhibited far lower DDH (<25%) and ANI (<81%) values, consistent with the current classification.
Outline of the procedure for detailed comparison of the chromosomes
The chromosome comparison strategy used here is the same as previously developed and applied to strains of P. laumondii (Zamora-Lagos et al., 2018). The sequence alignment program MAFFT (Katoh & Standley, 2013) was used to delineate matching segments (matchSEGs) which are common to both strains and divergent segments (divSEGs) which represent strain-specific genome regions (see the legend to Table 4 for details). In this way, genome sequences can be partitioned so that consecutive regions toggle between matchSEGs and divSEGs.
| nr | Class | Pos (91-R6) | Length (91-R6) | GC (%) | Pos (R1, shifted) | Pos (R1, original) | Length (R1) | GC (%) | Diff (%) | Difference details | Comment |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | matchSEG | 1–70,402 | 70,402 | 69.2 | 1–70,398 | 1,792,937–1,863,334 | 70,398 | 69.3 | 0.67 | 70,401/468/4/2 | – |
| 02 | indel | 70,403–70,535 | 133 | 57.1 | – | – | – | – | – | 133 bp deletion in R1 | |
| 02 | matchSEG | 70,536–183,138 | 112,603 | 67.9 | 70,399–183,020 | 1,863,335–1,975,956 | 112,622 | 67.9 | 0.63 | 112,601/708/6/29 | – |
| 03 | indel | – | – | 183,021–184,873 | 1,975,957–1,977,809 | 1,853 | 57.6 | – | – | MGE:ISH34 | |
| 03 | matchSEG | 183,139–217,136 | 33,998 | 65.5 | 184,874–218,882 | 1,977,810–10,856 | 34,009 | 65.7 | 1.96 | 33,976/655/12/67 | DissimLocal |
| 04 | replace | 217,137–264,198 | 47,062 | 56.3 | 218,883–280,477 | 10,857–72,451 | 61,595 | 56.1 | – | – | 91-R6:specific + MGE:multi; R1:specific + MGE:multi (AT-rich island) |
| 04 | matchSEG | 264,199–280,113 | 15,915 | 71.2 | 280,478–296,391 | 72,452–88,365 | 15,914 | 71.3 | 0.96 | 15,915/151/1/0 | – |
| 05 | replace | 280,114–281,650 | 1,537 | 53.4 | 296,392–305,571 | 88,366–97,545 | 9,180 | 59.1 | – | – | 91-R6:specific + MGE:HsTyIRS46; R1:specific + MGE:ISH8 |
| 05 | matchSEG | 281,651–284,185 | 2,535 | 65.2 | 305,572–308,111 | 97,546–100,085 | 2,540 | 65.0 | 1.81 | 2,536/45/1/4 | DissimGlobal |
| 06 | replace | 284,186–284,565 | 380 | 59.7 | 308,112–311,011 | 100,086–102,985 | 2,900 | 56.5 | – | – | 91-R6:specific; R1:specific |
| 06 | matchSEG | 284,566–299,230 | 14,665 | 66.6 | 311,012–325,676 | 102,986–117,650 | 14,665 | 66.6 | 0.12 | 14,665/17/0/0 | – |
| 07 | indel | – | – | 325,677–326,207 | 117,651–118,181 | 531 | 46.3 | – | – | MGE:ISH2 + TSD | |
| 07 | matchSEG | 299,231–400,107 | 100,877 | 70.4 | 326,208–427,087 | 118,182–219,061 | 100,880 | 70.4 | 0.10 | 100,876/97/3/8 | – |
| 08 | replace | 400,108–400,478 | 371 | 64.7 | 427,088–428,787 | 219,062–220,761 | 1,700 | 59.0 | – | – | 91-R6:specific; R1:MGE:ISH22 |
| 08 | matchSEG | 400,479–435,432 | 34,954 | 69.3 | 428,788–463,744 | 220,762–255,718 | 34,957 | 69.3 | 0.09 | 34,955/30/1/2 | – |
| 09 | indel | – | – | 463,745–465,157 | 255,719–257,131 | 1,413 | 58.5 | – | – | MGE:ISH8B + TSD | |
| 09 | matchSEG | 435,433–467,991 | 32,559 | 68.9 | 465,158–497,716 | 257,132–289,690 | 32,559 | 68.9 | 0.00 | 32,559/0/0/0 | – |
| 10 | indel | – | – | 497,717–498,846 | 289,691–290,820 | 1,130 | 58.8 | – | – | MGE:ISH1 + TSD | |
| 10 | matchSEG | 467,992–696,182 | 228,191 | 68.3 | 498,847–727,037 | 290,821–519,011 | 228,191 | 68.3 | 0.01 | 228,191/29/0/0 | – |
| 11 | replace | 696,183–696,402 | 220 | 68.2 | 727,038–727,082 | 519,012–519,056 | 45 | 62.2 | – | – | 91-R6:specific; R1:MGE:HsIRS31 |
| 11 | matchSEG | 696,403–796,274 | 99,872 | 68.9 | 727,083–826,948 | 519,057–618,922 | 99,866 | 68.9 | 0.60 | 99,869/597/4/4 | – |
| 12 | replace | 796,275–960,569 | 164,295 | 57.1 | 826,949–829,254 | 618,923–621,228 | 2,306 | 66.1 | – | – | 91-R6:specific + R1-plasmid-related + MGE:multi; R1:specific |
| 12 | matchSEG | 960,570–1,027,434 | 66,865 | 68.1 | 829,255–896,173 | 621,229–688,147 | 66,919 | 68.1 | 0.70 | 66,868/459/12/60 | – |
| 13 | indel | 1,027,435–1,028,828 | 1,394 | 58.7 | – | – | – | – | – | MGE:ISHsal1 + TSD | |
| 13 | matchSEG | 1,028,829–1,043,012 | 14,184 | 69.9 | 896,174–910,358 | 688,148–702,332 | 14,185 | 69.9 | 0.56 | 14,185/79/1/0 | – |
| 14 | indel | 1,043,013–1,046,256 | 3,244 | 49.4 | – | – | – | – | – | Specific | |
| 14 | matchSEG | 1,046,257–1,083,026 | 36,770 | 65.6 | 910,359–947,119 | 702,333–739,093 | 36,761 | 65.5 | 0.61 | 36,764/222/3/6 | – |
| 15 | indel | – | – | 947,120–956,649 | 739,094–748,623 | 9,530 | 54.1 | – | – | Specific | |
| 15 | matchSEG | 1,083,027–1,083,645 | 619 | 62.5 | 956,650–957,283 | 748,624–749,257 | 634 | 63.6 | 3.39 | 620/20/1/14 | DissimButShort |
| 16 | indel | – | – | 957,284–958,480 | 749,258–750,454 | 1,197 | 58.4 | – | – | Specific | |
| 16 | matchSEG | 1,083,646–1,084,546 | 901 | 60.4 | 958,481–959,381 | 750,455–751,355 | 901 | 59.9 | 3.00 | 901/27/0/0 | DissimButShort |
| 17 | replace | 1,084,547–1,087,761 | 3,215 | 49.0 | 959,382–960,467 | 751,356–752,441 | 1,086 | 49.3 | – | – | 91-R6:specific; R1:specific |
| 17 | matchSEG | 1,087,762–1,098,510 | 10,749 | 68.0 | 960,468–971,238 | 752,442–763,212 | 10,771 | 68.1 | 1.14 | 10,749/117/6/28 | DissimLocal |
| 18 | replace | 1,098,511–1,176,734 | 78,224 | 62.5 | 971,239–1,015,384 | 763,213–807,358 | 44,146 | 64.8 | – | – | 91-R6:specific; R1:specific + MGE:ISH6 |
| 18 | matchSEG | 1,176,735–1,203,631 | 26,897 | 69.6 | 1,015,385–1,042,275 | 807,359–834,249 | 26,891 | 69.6 | 0.56 | 26,892/149/1/5 | – |
| 19 | indel | 1,203,632–1,205,288 | 1,657 | 59.3 | – | – | – | – | – | MGE:HsTy_ISNpe8 + TSD | |
| 19 | matchSEG | 1,205,289–1,261,146 | 55,858 | 68.5 | 1,042,276–1,098,166 | 834,250–890,140 | 55,891 | 68.5 | 0.45 | 55,862/242/8/33 | – |
| 20 | indel | 1,261,147–1,263,140 | 1,994 | 47.2 | – | – | – | – | – | Specific | |
| 20 | matchSEG | 1,263,141–1,279,331 | 16,191 | 70.3 | 1,098,167–1,114,357 | 890,141–906,331 | 16,191 | 70.2 | 0.67 | 16,191/109/0/0 | – |
| 21 | indel | – | – | 1,114,358–1,116,307 | 906,332–908,281 | 1,950 | 58.6 | – | – | Specific | |
| 21 | matchSEG | 1,279,332–1,288,488 | 9,157 | 68.3 | 1,116,308–1,125,468 | 908,282–917,442 | 9,161 | 68.2 | 1.05 | 9,158/94/2/4 | DissimLocal |
| 22 | indel | 1,288,489–1,289,247 | 759 | 71.7 | – | – | – | – | – | Specific | |
| 22 | matchSEG | 1,289,248–1,289,304 | 57 | 66.7 | 1,125,469–1,125,525 | 917,443–917,499 | 57 | 70.2 | 3.51 | 57/2/0/0 | DissimButShort |
| 23 | indel | 1,289,305–1,289,550 | 246 | 73.6 | – | – | – | – | – | Specific | |
| 23 | matchSEG | 1,289,551–1,321,378 | 31,828 | 68.3 | 1,125,526–1,157,344 | 917,500–949,318 | 31,819 | 68.5 | 1.03 | 31,822/324/4/7 | DissimLocal |
| 24 | indel | 1,321,379–1,323,383 | 2,005 | 53.7 | – | – | – | – | – | Specific | |
| 24 | matchSEG | 1,323,384–1,352,409 | 29,026 | 69.3 | 1,157,345–1,186,375 | 949,319–978,349 | 29,031 | 69.2 | 0.34 | 29,027/99/1/4 | – |
| 25 | indel | – | – | 1,186,376–1,187,505 | 978,350–979,479 | 1,130 | 59.0 | – | – | MGE:ISH1 + TSD | |
| 25 | matchSEG | 1,352,410–1,408,011 | 55,602 | 69.8 | 1,187,506–1,243,108 | 979,480–1,035,082 | 55,603 | 69.8 | 0.22 | 55,603/119/1/0 | – |
| 26 | indel | 1,408,012–1,408,424 | 413 | 48.7 | – | – | – | – | – | MGE:MITEHsal2 + TSD | |
| 26 | matchSEG | 1,408,425–1,455,590 | 47,166 | 69.6 | 1,243,109–1,290,274 | 1,035,083–1,082,248 | 47,166 | 69.6 | 0.00 | 47,166/1/0/0 | – |
| 27 | indel | 1,455,591–1,456,001 | 411 | 47.7 | – | – | – | – | – | MGE:MITEHsal2 + TSD | |
| 27 | matchSEG | 1,456,002–1,593,555 | 137,554 | 68.6 | 1,290,275–1,427,889 | 1,082,249–1,219,863 | 137,615 | 68.6 | 0.18 | 137,557/251/3/58 | – |
| 28 | indel | 1,593,556–1,595,147 | 1,592 | 58.6 | – | – | – | – | – | MGE:ISH10 + TSD | |
| 28 | matchSEG | 1,595,148–1,595,381 | 234 | 46.2 | 1,427,890–1,428,123 | 1,219,864–1,220,097 | 234 | 46.2 | 0.00 | 234/0/0/0 | – |
| 29 | indel | – | – | 1,428,124–1,428,655 | 1,220,098–1,220,629 | 532 | 45.9 | – | – | MGE:ISH2 + TSD | |
| 29 | matchSEG | 1,595,382–1,621,874 | 26,493 | 66.7 | 1,428,656–1,455,148 | 1,220,630–1,247,122 | 26,493 | 66.7 | 0.00 | 26,493/1/0/0 | – |
| 30 | indel | 1,621,875–1,629,435 | 7,561 | 52.7 | – | – | – | – | – | Specific | |
| 30 | matchSEG | 1,629,436–1,734,188 | 104,753 | 68.0 | 1,455,149–1,559,901 | 1,247,123–1,351,875 | 104,753 | 68.0 | 0.01 | 104,753/14/0/0 | – |
| 31 | indel | 1,734,189–1,739,027 | 4,839 | 59.3 | – | – | – | – | – | Specific | |
| 31 | matchSEG | 1,739,028–1,972,467 | 233,440 | 69.1 | 1,559,902–1,793,340 | 1,351,876–1,585,314 | 233,439 | 69.1 | 0.13 | 233,439/311/3/4 | – |
| 32 | indel | 1,972,468–1,973,942 | 1,475 | 70.4 | – | – | – | – | – | Specific | |
| 32 | matchSEG | 1,973,943–1,985,336 | 11,394 | 71.2 | 1,793,341–1,804,733 | 1,585,315–1,596,707 | 11,393 | 71.2 | 0.90 | 11,394/102/1/0 | – |
| 33 | replace | 1,985,337–1,986,883 | 1,547 | 56.6 | 1,804,734–1,806,683 | 1,596,708–1,598,657 | 1,950 | – | – | 91-R6:specific; R1:specific | |
| 33 | matchSEG | 1,986,884–2,010,338 | 23,455 | 69.2 | 1,806,684–1,830,134 | 1,598,658–1,622,108 | 23,451 | 69.4 | 0.82 | 23,454/186/6/4 | – |
| 34 | indel | 2,010,339–2,010,923 | 585 | 50.4 | – | – | – | 46.2 | – | – | Specific |
| 34 | matchSEG | 2,010,924–2,040,807 | 29,884 | 69.5 | 1,830,135–1,860,018 | 1,622,109–1,651,992 | 29,884 | 69.5 | 0.13 | 29,884/38/0/0 | – |
| 35 | indel | 2,040,808–2,041,738 | 931 | 67.8 | – | – | – | – | – | Specific | |
| 35 | matchSEG | 2,041,739–2,042,856 | 1,118 | 67.6 | 1,860,019–1,861,136 | 1,651,993–1,653,110 | 1,118 | 68.0 | 2.77 | 1,118/31/0/0 | DissimButShort |
| 36 | indel | 2,042,857–2,043,910 | 1,054 | 70.0 | – | – | – | – | – | Specific | |
| 36 | matchSEG | 2,043,911–2,093,804 | 49,894 | 70.0 | 1,861,137–1,911,041 | 1,653,111–1,703,015 | 49,905 | 70.1 | 0.73 | 49,894/361/4/15 | – |
| 37 | indel | 2,093,805–2,094,695 | 891 | 67.6 | – | – | – | – | – | Specific | |
| 37 | matchSEG | 2,094,696–2,113,373 | 18,678 | 70.1 | 1,911,042–1,929,720 | 1,703,016–1,721,694 | 18,679 | 70.3 | 0.56 | 18,679/103/1/0 | – |
| 38 | replace | 2,113,374–2,113,457 | 84 | 72.6 | 1,929,721–1,930,496 | 1,721,695–1,722,470 | 776 | 70.4 | – | – | 91-R6:specific; R1:specific |
| 38 | matchSEG | 2,113,458–2,165,583 | 52,126 | 68.0 | 1,930,497–1,982,626 | 1,722,471–1,774,600 | 52,130 | 68.0 | 0.72 | 52,128/372/4/4 | – |
| 39 | indel | – | – | 1,982,627–1,987,937 | 1,774,601–1,779,911 | 5,311 | 66.0 | – | – | Specific | |
| 39 | matchSEG | 2,165,584–2,178,608 | 13,025 | 69.2 | 1,987,938–2,000,962 | 1,779,912–1,792,936 | 13,025 | 69.3 | 0.68 | 13,025/89/0/0 | – |
Note
- The chromosomes toggle between matching segments (matchSEG) and divergent segments (divSEG). MatchSEGs have an assigned serial number (nr). Each divSEG is labeled by the serial number of the subsequent matchSEG. DivSEGs are classified (class) as indel or replace. Indels are assigned with one-base resolution and are either an insertion in one strain or a deletion in the other. In the case of a replacement (“replace”), the chromosomal alignment is interrupted by independent strain-specific sequences. These may be completely unrelated but may also be homologous (but below 85% DNA sequence identity). For both strains, the length (length) and the position (pos) of the segment is given. For strain R1, the position of the original sequence (GenBank:AM774415) is provided, as well as the position for a shifted sequence where the point of ring opening is adjusted to that for strain 91-R6. MatchSEG03 traverses the point of ring opening of the original sequence and covers bases 1,977,810–2,000,962 and 1–10,856 (“1,977,810–10,856”). For all segments, the GC content is given (“GC (%)”). Low-GC MGEs have a gray background color, other low-GC divSEGs are highlighted in yellow, high-GC divSEGs in green. For matchSEGs, the sequence difference (diff) is typically below 1%. For those with >1% sequence difference, an explanation is provided (comment). DissimButShort points to short matchSEGs (max 1,118 bp, up to 3.51% sequence difference). DissimGlobal points to an enhanced divergence over the major part of the matchSEG (one case; matchSEG length 2,540 bp, 1.81% sequence difference). DissimLocal points to a restricted region with enhanced divergence (max 1.96% sequence difference). Details (difference details) are a series of four integers separated by slash. They refer to the number of matching bases, mismatching bases, gap open characters, and gap extension characters, respectively. The comment (comment) also provides some detail for divSEGs. In case of a replacement, terms are provided for each of the strains, 91-R6 and R1. For MGE (mobile genetic element), the type of transposon (ISH) or MITE is indicated. MGEs may be associated with a target site duplication (+TSD). The term MGE:multi refers to multiple MGEs on the divSEG. The term “specific” is used when the divSEG sequence is strain-specific and thus not (or only distantly) related to the sequence from the other strain.
Matching genome segments between the chromosomes of strain 91-R6 and strain R1
Alignment of the chromosomes of strains 91-R6 and R1 revealed they are highly similar and completely colinear (Figure 1, Table 4), with an overall sequence identity of 99.63%. There are 39 matching segments (matchSEGs) that together cover the majority of both chromosomes (1.85 Mb; 84.9% for the Hbt. salinarum strain 91-R6 chromosome and 92.5% for strain R1), and between these are 38 strain-specific sequences. Thirty of the 39 matchSEGs show <1% sequence divergence, while the remaining nine matchSEGs have more than 1% sequence difference (average 1.47%) but are relatively short (90 kb total). Overall, 6,719 point mutations and 87 small indels were detected in the 39 matchSEGs (65 indels < 20 nt, longest indel 79 nt).
Strain-specific regions in the chromosomes of strains 91-R6 and R1
The strain-specific sequences (referred to as divergent segments, divSEGs) sum up to 328,119 bp for Hbt. salinarum strain 91-R6 (15.1% of its genome) and 150,261 bp for strain R1 (7.5% of its genome).
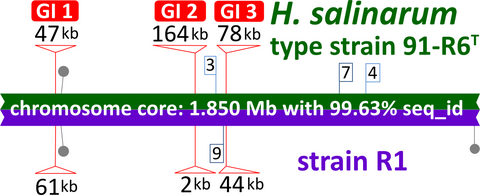
DivSEGs were classified into two categories, indels and replacements (Table 4). Indels refer to sequences that are contiguous in one genome while the other has an insertion of additional sequence, and where this insertion can be pinpointed to an exact position. There are 18 insertions in strain 91-R6 and 10 insertions in strain R1. Several of the insertions are MGEs, which are described in more detail below (Section 3.3). Replacements are where the two strains have dissimilar sequences located at an equivalent position, and the borders can be discerned with 1-base resolution. A total of 10 replacements were detected. Most of the sequences in replacements were completely unrelated between the strains. For sequence regions longer than 1 kb, we found an upper limit of 80% DNA sequence identity, which indicates independent sequences in a genome context with >99% DNA sequence identity. The locations of most divSEGs are visible in Figure 1 as white gaps in the BLASTn ring of the chromosome. Only three divSEGs exceed 10 kb, and these represent the three large GIs detected by Island Viewer (see below and Figure 1).
Correlation of protein-coding genes among the three analyzed strains
As an annotation principle, every gene encoding a protein on a matchSEG in one strain must have a correlated gene in the other strain. The gene sets of the three strains have been correlated in detail (see Section 2 and Appendix 4 and Appendix 5). Proteins are classified as strain-specific only after validation by tBLASTn that they are not mere missing gene calls. Correlated proteins encoded on the chromosome of strains 91-R6 and R1 are listed in Table S1 (1,986 proteins) (via Zenodo; https://doi.org/10.5281/zenodo.3528126). The corresponding proteins from strain NRC-1 are also listed. In addition, there are regions of very high similarity between the plasmids from strains 91-R6 and R1 (see below), and the resulting plasmid-encoded correlated proteins are listed in Table S2 (via Zenodo). Furthermore, Table S2 lists proteins which are encoded on a plasmid in strain R1 but in a strain-specific region of the chromosome from strain 91-R6 (see below). Chromosomally encoded proteins from strain-specific regions of 91-R6 are listed in Table S3 (via Zenodo). In several cases, a homolog exists in R1, but the genes are not positionally correlated. Such homologs are also listed in Table S3. Residual proteins which are specific for the chromosome of strain R1 are listed in Table S4 (via Zenodo). Plasmid-encoded proteins specific for strain 91-R6 are listed in Table S5 (via Zenodo), while plasmid-encoded proteins specific for strain R1 are listed in Table S6 (via Zenodo). Some of the strain-specific plasmid proteins are discussed in more detail (see below). A few protein-coding genes exist in strain NRC-1 but are absent in both strains 91-R6 and R1. These are listed in Table S7 (via Zenodo). Finally, Table S8 (via Zenodo) lists ORFs which are annotated in the current version of the NRC-1 genome (AE004437, AE004438, AF016485) but which are considered not to code for a protein (spurious ORFs; for definition, see Appendix 4).
Proteins encoded on strain-specific chromosomal regions
The characteristics of the longest divSEGs are described here. For an analysis of other divSEGs see Appendix 7. The three longest divSEGs (04, 12, 18) are GIs (GI-1, GI-2, GI-3) (as indicated in Figure 1).
DivSEG04 is a replacement where the R1 sequence is 61,595 bp long and represents the well-known “AT-rich island” (GC content reduced to 56.1%) (Joshi, Guild, & Handler, 1963; Moore & McCarthy, 1969; Ng et al., 2000; Pfeifer & Betlach, 1985). At the equivalent position in strain 91-R6 is a 47,062 bp region, which also has a reduced GC content (56.3%), and both are likely to represent horizontally transferred DNA. Both regions carry many mobile genetic elements, at least one Orc paralog, and are rich in glycosyltransferases and other sugar metabolism related genes. The DNA sequences are mostly unrelated (only two BLASTn matches exceed 1 kb), but eight encoded proteins are homologous and show up to 86% protein sequence identity. Nearby and upstream of this region are genes for the S-layer glycoprotein (HBSAL_01075), secreted glycoproteins (HBSAL_01070, 01065), and sugar nucleotidyltransferases (HBSAL_01110, 01105), and a potential role for this replacement region is to provide an altered repertoire of sugars for modifying secreted glycoproteins (e.g. S-layer) and extracellular polysaccharides (EPS), perhaps to avoid virus predation. We propose that this replacement region be called genomic island 1 (GI-1) (Figure 1).
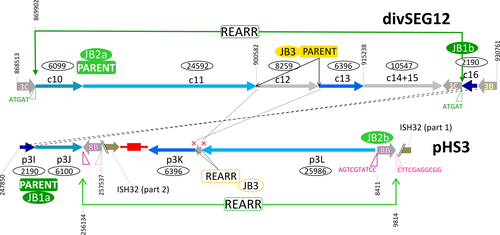
DivSEG12, which corresponds to genomic island GI-2, is the longest strain-specific sequence in strain 91-R6 (164,295 bp). In R1, there is a 2,306 bp region at the same genome position. The R1 sequence codes for most of the alpha subunit of dimethylsulfoxide reductase (dmsA, codons 69–836 of 837), while the N-terminal 68 residues are encoded on the preceding matchSEG. The termini of the 164,295 bp region in strain 91-R6 code for a close homolog of DmsA (57% protein sequence identity) which has been disrupted due to targeting by MITEHsal3. The integration point corresponds to codon 622 of R1 DmsA. The concatenated protein sequence was most similar (78% amino acid identity; BLASTp) to a homolog from Halostella sp. DLLS-108 (accession WP_135820841.1). The long N-terminal fragment (HBSAL_04215) covers the 4Fe-4S (IPR006963) and catalytic (IPR006656) domains. The C-terminal fragment (HBSAL_05135) covers the molybdopterin dinucleotide-binding domain (IPR006657). The 91-R6 specific 164 kb region has a GC content below 60% and carries several Orc paralogs and multiple MGEs, thus showing characteristics of an integrated plasmid. The full copy of a MITEHsal3 at one end and a partial copy (truncated due to MGE targeting) at the other suggests that, after the initial MITE insertion into dmsA, there were further integration events that initially targeted the MITE. This is further supported by a hybrid TSD (TATGACA) around these copies of MTEHsal3. Among the proteins encoded in the 91-R6 specific region are multiple paralogs of TATA-binding transcription factors. Several of the encoded proteins are close homologs of proteins encoded on the plasmids from strain R1 (see below and Table S2 (via Zenodo)). Four sequences, totaling 42.5 kb, show a close but complex relationship to R1 plasmid pHS3 (see below, Figure 2, and Tables 5 and 6).
| Tag | Pos (R1; pHS3) | Length (R1) | GC (%) | Sequence comparison | Pos (91-R6) | Length (91-R6) | Pos (NRC-1) | Length (NRC-1) | Comment |
|---|---|---|---|---|---|---|---|---|---|
| 8,412–9,814 | 1,403 | – | – | – | – |
pNRC200 fwd 122,259–123,661 |
1,403 | MGE:ISH8B | |
| 9,815–11,315 | 1,501 | – | – | – | – |
pNRC200 fwd 123,662–125,162 |
1,501 | MGE:ISH32 (part 1; rev) | |
| p3A | 11,316–42,736 | 31,421 | 56.3 | – | – | – |
pNRC200 fwd 125,163–159,391 |
34,239 | MGE:multi; NRC-1 targeted by ISH3 + ISH8; the R1 and NRC-1 sequences are identical except for 7-point mutations within an ISH3 |
| pp11-pp14 | 42,737–150,574 | 107,838 | – | – |
pHSAL1 fwd 1–109,176 |
109,176 |
pNRC200 fwd 159,392–265,437 |
106,046 | Extensive match between pHSAL1 and pHS3 (see Table 9) |
| p3F | 150,575–244,107 | 93,533 | 58.5 | – | – | – | – | – | MGE:multi |
| p3G | 244,108–247,091 | 2,983 | 58.8 | Identical |
pHSAL1 rev 144,795–141,813 |
2,983 | – | – | dup(pHSAL1 + 2) |
| 247,092–247,612 | 521 | – | – | – | – | – | – | MGE:ISH2 + TSD (TSD 55 nt) | |
| p3H | 247,613–247,854 | 242 | 50.0 | Identical |
pHSAL1 rev 141,867–140,222 |
1,646 | – | – | dup(pHSAL1 + 2); targeted by ISHsal2 (91-R6); at3pr: MGE:ISH3B |
| p3I | 247,850–250,039 | 2,190 | 52.6 | Identical |
Chr rev 929,363–927,174 |
2,190 | – | – | at5pr: MGE:ISH3B; at3pr: MGE:ISH3C |
| p3J | 250,035–256,134 | 6,100 | 57.3 | 1 Point mutation |
Chr fwd 869,902–876,000 |
6,099 | – | – | at5pr: MGE:ISH3C |
| 256,135–257,537 | 1,403 | – | – | – | – | – | – | MGE:ISH8B | |
| 257,538–259,840 | 135 | – | – | – | – | – | – | MGE: ISH32 (part 2; fwd) | |
| 256,135–259,840 | 2,168 | – | – | – | – | – | – | Specific; MGE:multi | |
| p3K | 259,840–266,236 | 6,396 | 59.8 | 1 Point mutation |
Chr rev 915,237–908,842 |
6,396 | – | – | – |
| 266,237–266,757 | 521 | – | – | – | – | – | – | MGE:ISH2 (no TSD) | |
| p3L | 266,758–8,411 | 25,986 | 61.0 | Identical |
Chr rev 900,582–875,991 |
24,592 | pNRC100 rev 150,253–133,743; pNRC200 fwd 113,317–121,727 | 16,511; 8,411 | MGE:multi; targeted by ISH3(R1, NRC-1); rearranged(NRC-1) |
Note
- Plasmid pHS3 is a patchwork of unique and shared sequences. Region tags start with p3 (plasmid pHS3 regions) (tag), except for those matching to the unique part of pHSAL1 (pp11–pp14, see Table 9). This match starts at pHS3 position 42,737, the sequence being shared in forward orientation (“pHSAL1 fwd”) (column pos). The preceding 31.4 kb region p3A is not present in strain 91-R6 but is shared and contiguous with pNRC200 from NRC-1 (including the upstream MGEs). The subsequent 93 kb region p3F is unique to strain R1. Within this region, several MGEs are found (“MGE:multi”). The next set of regions is found in strain 91-R6 but is missing from strain NRC-1. Regions p3G and p3H match to a region from the 39 kb duplication between plasmids pHSAL1 and pHSAL2 (“dup(pHSAL1 + 2)”), being located on the reverse strand (“pHSAL1 rev”). In pHS3, these regions are separated by a copy of ISH2 which is positioned within an exceedingly long TSD (“TSD 55 nt”). The sequences (3.2 kb total) are identical (“identical” in column “sequence comparison”) except for MGE targeting in strain 91-R6 (“targeted by ISHsal2 (91-R6)”). In strain 91-R6, region p3H terminates at its 3′ end with an ISH3 of subtype ISH3B (“at3pr: MGE:ISH3B”). While this sequence is contiguous with region p3I in pHS3, the matching region in strain 91-R6 is found in the chromosome in reverse orientation (“Chr rev”) where it is part of divSEG12 (Table 6; tag c16; see Figures 2 and 3). This region also starts with ISH3B. Regions p3H and p3I overlap by five bases (AAATT) which thus are reminiscent of a TSD even though the copies of ISH3B are on distinct replicons in strain 91-R6. An equivalent situation exists between regions p3I and p3J which are contiguous in pHS3, terminate with equivalent MGEs (subtype ISH3C) in strain 91-R6, and overlap by five bases (ATGAT). In this case, both regions are in divSEG12 in strain 91-R6, but at distinct locations and in opposite orientation (Table 6; tags c10 and c16). Separated by a MGE conglomerate is region p3K which again is part of divSEG12 but also positionally disconnected. Junctional analysis identified a genome inversion in pHS3 (disconnects p3J from p3L; Figure 6). Regions p3K and p3L are 8 kb apart in strain 91-R6 but separated by only a single ISH2 on pHS3. This MGE is not bounded by a TSD (“no TSD”), which thus may indicate an ISH2-triggered deletion in strain R1. Region p3L is shared between all three strains with complete sequence identity except for one targeting MGE in R1 (which also is found in strain NRC-1; “targeted by ISH3(R1,NRC-1)”). Otherwise, the sequences are colinear between strains 91-R6 and R1, while the sequence is rearranged in strain NRC-1 (“rearranged(NRC-1)”). Part of this sequence (8,411 bp) is found on pNRC200, as are most of the sequences shared between strain NRC-1 and R1 plasmid pHS3. The central region (1,065 bp) is absent from strain NRC-1. The remainder (16,511 bp) has been shifted to plasmid pNRC100 in reverse orientation. Region p3L traverses the point of ring opening of pHS3 (266,758–284,332;1–8,411 represented as “266,758–8,411”) the setting of which has been based on the match between pHS3 and pNRC200. In the 42.5 kb which are shared between strains 91-R6 and R1 in total, only two point mutations are encountered. Likely events which split the 42.5 kb shared sequence into four regions (p3I, p3J, p3K, and p3L) are described in the text. Junctions are described in Appendix 8.
| Tag | Pos (91-R6; Chr) | Length (91-R6) | GC (%) | Sequence comparison | Pos (R1; plasmid) | Length (R1) | Pos (NRC-1; plasmid) | Length (NRC-1) | Comment |
|---|---|---|---|---|---|---|---|---|---|
| c01 | 796,275–800,761 | 4,487 | 56.7 | – | – | – | – | – | Unique + MGE:MITEHsal3 |
| c02 | 800,762–804,395 | 3,634 | 58.8 | 95% |
pHS2 rev 25,805–30,927 |
5,123 |
pNRC200 rev 294,049–299,171 |
5,123 | Targeted by MGE:ISH20 (R1, NRC-1) |
| c03 | 804,396–834,825 | 30,430 | 55.6 | – | – | – | – | – | Unique + MGE:multi |
| c04 | 834,826–835,650 | 825 | 53.3 | 100% |
pHS1 fwd 142,495–143,319 |
825 |
pNRC100 fwd 129,104–129,928 |
825 | dup(pHS1 + 4) |
| c05 | 835,651–853,415 | 17,765 | 55.9 | – | – | – | – | – | Unique + MGE:multi |
| c06 | 853,416–854,070 | 655 | 57.7 | 97% |
pHS2 fwd 60,973–62,158 |
1,186 |
pNRC200 fwd 329,748–330,402 |
655 | Targeted by MGE:ISH2 (R1) |
| 854,071–854,475 | 405 | – | – | – | – | – | – | MGE:MITEHsal2 | |
| c07 | 854,476–854,608 | 133 | 45.9 | 100% | pHS2 fwd 62,303–62,435 | 133 |
pNRC200 fwd 330,547–330,679 |
133 | – |
| c08 | 854,606–856,247 | 1,642 | 56.9 | 99% |
pHS1 rev 59,298–60,939 |
1,642 |
pNRC100 rev 58,449–61,503 |
3,055 | dup(pHS1 + 2); dup(pNRC100 + 200); targeted by MGE:ISH8 (NRC-1) |
| c09 | 856,248–868,512 | 12,265 | 54.4 | – | – | – | – | – | Unique + MGE:multi |
| 868,513–869,901 | 1,389 | – | – | – | – | – | – | MGE:ISH3 | |
| c10 | 869,902–876,000 | 6,099 | 57.3 | 99% (1 one-base indel) |
pHS3 fwd 250,035–256,134 |
6,100 | – | – | – |
| c11 | 875,991–900,582 | 24,592 | 61.0 | 100% |
pHS3 rev 266,758–284,332; 1–8,411 |
25,986 |
pNRC200 rev 113,317–121,727; pNRC100 fwd 133,743–150,253 |
8,411; 16,511 | MGE:multi; targeted by MGE:ISH3D (R1, NRC-1); rearranged(NRC-1) |
| c12 | 900,583–908,841 | 8,259 | 56.7 | – | – | – | – | – | Unique + MGE:multi |
| c13 | 908,842–915,237 | 6,396 | 59.7 | 99% (1 point mutation) |
pHS3 rev 259,841–266,236 |
6,396 | – | – | – |
| c14 | 915,238–921,275 | 6,038 | 62.9 | 87% |
pHS1 rev 45,663–54,529 |
8,867 |
pNRC100 rev 44,814–53,680 |
8,867 | Dup(pHS1 + 2); Dup(pNRC100 + 200); 3 kb deletion in 91-R6 |
| c15 | 921,276–925,784 | 4,509 | 62.8 | ||||||
| 925,785–927,173 | 1,389 | – | – | – | – | – | – | MGE:ISH3 | |
| c16 | 927,174–929,363 | 2,190 | 52.6 | 100% |
pHS3 rev 247,850–250,039 |
2,190 | – | ||
| 929,364–930,761 | 1,398 | – | – | – | – | – | – | MGE:ISH3 | |
| c17 | 930,762–960,569 | 29,808 | 54.9 | – | – | – | – | – | Unique + MGE:multi |
Note
- DivSEG12 was split into subregions based on BLASTn results (excluding MGE sequences from BLASTn analysis). Junctions were chosen to reflect beginning/end of close matches to strain R1 (and NRC-1). Region tags start with c (chromosomal) (tag). The comment column (column) term “unique” indicates the absence of a close homolog (>90% DNA sequence identity) in R1. When the region carries MGEs, this is indicated (single MGEs are named; otherwise “MGE:multi”). For all matches, the plasmid is indicated. It is also indicated if the match is to the forward (“fwd”) or reverse (“rev”) strand. If the homologous region is on a plasmid duplication in R1, the position is given for pHS1 and the duplication is indicated in the comment column (“dup(pHS1 + 2)” or “dup(pHS1 + 4)”). If the homologous region is on a plasmid duplication in NRC-1, the position applies to both, pNRC100 and pNRC200, and the duplication is indicated in the comment column (“dup(pNRC100 + 200”). We do not specify potential additional copies on inverted duplications. If a match carries an additional, strain-specific MGE, the MGE is named and the affected strains are listed (e.g. “targeted by MGE:ISH20 (R1,NRC-1)”). MGEs which are specific for strain 91-R6 are listed without region tag. For none of the 91-R6 specific MGEs was a TSD encountered, which may indicate postintegration genome rearrangements. In some cases, adjacent subregions match to disconnected regions on the R1 plasmids (c07/c08; c10/c11; c13/c14). For the meaning of “rearranged(NRC-1)” see the legend to Table 5. In subregion c14, strain 91-R6 suffered a 3 kb deletion so that two genes are absent and one is truncated (HBSAL_04945) (“3 kb deletion in 91-R6”).
DivSEG18 corresponds to a replacement where the R1 sequence is 44,146 bp long and the 91-R6 sequence at the equivalent genome position has 78,224 bp. This corresponds to genomic island GI-3. While both sequences have more than 60% GC, it is slightly reduced from the genome average due to the presence of several MGEs. The region from strain 91-R6 contains several transposons which are specific for this strain (canonical transposons ISHsal1, ISHsal2, ISNpe16, and HsIRS45; noncanonical transposons ISHsal5, ISHsal12, and ISHsal14; see below, Section 3.3, Table 7 and Appendix 11). The 2nd ORF in the R1 region is an integrase domain protein, and it may be no accident that just upstream of divSEG18 is a tRNA-Met gene (a typical arrangement for integrative elements). At the genome level, the two sequences show restricted sequence similarity (up to 80% DNA sequence identity for regions up to 3 kb). Although these sequences are strain-specific, they code for distant homologs, and even retain partial gene synteny. In this context, homologs are considered distant even at 85% protein sequence identity, as orthologs within matchSEGs show at least 98% protein sequence identity (with very few exceptions). Remarkably, the only gene copies of triosephosphate isomerase (tpiA), histidinol-phosphate aminotransferase (hisC), and archaetidylglycerolphosphate synthase (agsA) are encoded on divSEG18. Also encoded is GTP cyclohydrolase 3 (IIa), which is involved in riboflavin biosynthesis (arfA1 in both strains, an additional arfA2 paralog only in strain R1). DNA polymerase Y is encoded in this region, but is disrupted in strain 91-R6. Some proteins which are physiologically relevant are encoded by more than one paralogous gene, of which one is located on divSEG18, including a probable adenylate kinase (adk2) and arlA, the gene coding for one of the archaellins (see above, Section 3.1). In R1, htr13, one of the methyl-accepting chemotaxis proteins (haloarchaeal transducers), is encoded close to the arlA locus while a transducer is not encoded on divSEG18 in strain 91-R6. Several enzymes of the N-glycosylation pathway are encoded on divSEG18 (see above, Section 3.1). The last bases of divSEG18 code for the N-terminal 18 codons of aglB, while the remainder of the protein is encoded on the subsequent matchSEG.
| Class | Name | MGE type | Count(R1) | Count(NRC-1) | Count(91-R6) | Occurrence | Targeting activity | Potential source |
|---|---|---|---|---|---|---|---|---|
| ISH3 | ISH3B | TNP | 3 | 3 | 2 | Common | – | – |
| ISH3 | ISH3C | TNP | 9 | 11 | 4 | Common | – | – |
| ISH3 | ISH3D | TNP | 2 | 2 | 1 | Common | – | – |
| ISH3 | ISH20 | TNP | 1 | 1 | – | Lab | – | R:pHS2 |
| ISH3 | ISHsal1 | TNP | – | – | 5 | Type | T:1(d13) | T:d12;d18;pHSAL2(p208) |
| ISH3 | ISHsal2 | TNP | – | – | 2 | Type | T:1(p206/p207) | T:d18 |
| ISH4 | ISH4 | TNP | 3 | 2 | – | Lab | – | R:pHS1 |
| ISH4 | ISHsal15 | TNP | – | – | 1 | Type | – | T:d12 |
| ISH4 | MITEHsal1 | MITE | 1 | 1 | 1 | Common | – | – |
| ISH4 | MITEHsal12 | MITE | – | – | 1 | Type | – | T:d12 |
| ISH6 | ISH6 | TNP | 3 | 2 | 1 | Common | – | – |
| ISH8 | ISH2 | MITE | 11 | 10 | – | Lab | R:4(d07,d29,p3G/H,p3K/L13) | R:pHS1;pHS2 |
| ISH8 | ISH5 | TNP | 1 | 1 | 2 | Common | – | – |
| ISH8 | ISH8A | TNP | 3 | 1 | 3 | Common | – | – |
| ISH8 | ISH8B | TNP | 9 | 6 | 1 | Common | R:1(d09) | – |
| ISH8 | ISH8C | TNP | 1 | 1 | 1 | Common | – | – |
| ISH8 | ISH8D | TNP | 3 | 3 | – | Lab | – | R:d04;pHS1;pHS2 |
| ISH8 | ISH8E | TNP | 1 | 4 | – | Lab | – | R:pHS1 |
| ISH8 | ISH30 | TNP | 1 | 1 | – | Lab | R:1(pp13) | R:unknown |
| ISH8 | ISH32 | TNP | 1 | (1) | 2 | Common | – | – |
| ISH8 | ISHsal3 | TNP | – | – | 1 | Type | – | T:d12 |
| ISH8 | ISHsal4 | TNP | – | – | 2 | Type | – | T:pHSAL2(p204,ppd) |
| ISH8 | MITEHsal6 | MITE | – | – | 1 | Type | – | T:d12 |
| ISH9 | ISH1 | TNP | 4 | 1 | 2 | Common | R:2(d10,d25) | – |
| ISH9 | ISH9 | TNP | 1 | 1 | 1 | Common | – | – |
| ISH9 | ISHsal6 | TNP | – | – | 1 | Type | – | T:unknown |
| ISH9 | HsIRS49 | TNP | – | – | 1 | Type | – | T:d12 |
| ISH9 | MITEHsal7 | MITE | 1 | 1 | – | Lab | – | R:d04 |
| ISH9 | MITEHsal13 | MITE | – | – | 1 | Type | – | T:d04 |
| ISH10 | ISH10 | TNP | 4 | 2 | 1 | Common | T:1(d28) | – |
| ISH10 | ISHsal7 | TNP | – | – | 1 | Type | – | T:d04 |
| ISH10 | ISNpe8 | TNP | 1 | 1 | 2 | Common | T:2(d19,pp12/pp13) | – |
| ISH11 | ISH11 | TNP | 1 | 4 | – | Lab | – | R:pHS2 |
| ISH11 | ISHsal8 | TNP | – | – | 1 | Type | – | R:pHSAL2(p208) |
| ISH11 | ISNpe16 | TNP | – | – | 2 | Type | – | T:d12,d18 |
| ISH11 | MITEHsal2 | MITE | – | – | 8 | Type | T:2(d26,d27) | – |
| ISH11 | MITEHsal3 | MITE | – | – | 2 | Type | T:1(pp11/pp12) | T:d12 |
| ISH11 | MITEHsal11 | MITE | 1 | 1 | 1 | Common | – | – |
| ISH11 | MITEHsal14 | MITE | – | – | 1 | Type | – | T:d12 |
| ISH14 | ISH29 | TNP | 1 | 1 | – | Lab | – | R:pHS2 |
| ISH14 | HsIRS45 | TNP | – | – | 1 | Type | – | T:d18 |
| ISH16 | ISHsal16 | TNP | – | – | 1 | Type | – | T:d12 |
| ISH16 | HsIRS12 | TNP | 1 | 1 | – | Lab | – | R:d04 |
| ISHwal16 | ISHsal9 | TNP | – | – | 1 | Type | – | T:d12 |
| ISHwal16 | ISHsal10 | TNP | – | – | 1 | Type | – | T:d12 |
| ISHwal16 | ISHsal11 | TNP | – | – | 1 | Type | – | T:d04 |
Note
- Two types of MGEs are considered (“MGE type”), transposons (“TNP”) and MITEs (“MITE”) (see Appendix 11 for definitions). MGEs are classified and named (“name”) according to ISFinder (Siguier et al., 2012). For atypical names see Appendix 11. MGEs with homologous transposase genes (or, in case of MITEs, homologous termini) are assigned to the same class (“class”). Only canonical MGEs (i.e. those with inverted terminal repeats) are considered and only complete copies are counted (for details see Appendix 11). In our definition, a complete MGE copy has both termini intact and is devoid of long internal deletions but may have been targeted by another MGE. We count (“count()”) the number of complete copies for each MGE in the three strains which are under study. The ISH32 is NRC-1 is equivalent to that in R1, the fragments of which have been disconnected by a genome inversion; in NRC-1, only the fragment upstream of p3A (see Table 5) is retained while the other part was lost by a strain-specific deletion. MGEs may occur in all three strains (“common”), only in the type strain 91-R6 (“type”), or only in the laboratory strains R1 and NRC-1 (“lab”). Strain-specific MGE copies occurring in a conserved genomic context indicate genome targeting (“targeting activity”). For strains 91-R6 (“T”) and R1 (“R”), the number of such targeting events and their location is provided. The term “T:1(d13)” indicates one targeting event in strain 91-R6 which is recorded in Table 4 as divSEG13. The term “T:1(p206/p207)” reflects an event that is recorded in Table 8 between regions p206 and p207. Likewise, the term “T:1(pp11/pp12)” points to the element in Table 9 between pp11 and pp12. We attempted to identify the potential source of MGEs which are specific for the type strain or the laboratory strains (“potential source”) (for details see Appendix 11). As such, the long replacement regions (divSEG04, divSEG12, divSEG18) or the plasmids were identified. For plasmids from strain 91-R6 and pHS3 from strain R1, the region tag from the appropriate table is also included. In ambiguous cases, multiple potential sources are listed. The term “unknown” indicates that the persisting copies have targeted another transposon in a region that is not strain-specific and accordingly a potential source cannot be discerned.
Other divSEGs which are briefly described in Appendix 7 are divSEG05, the divSEG22/23 pair, divSEG27, divSEG32, divSEG37, and divSEG39. Several divSEGs code for integrase domain proteins, and some of those have a tRNA gene at or close to the integration point (an arrangement typical of integrative mobile elements). This applies to the divSEG15/16/17 trio and divSEG30. DivSEG30 looks suspiciously like a provirus (7.5 kb long; targets a tRNA; one integrase family gene; 10 other genes, none of them well characterized) but does not match any virus in GenBank. Notably, divSEG14 and divSEG31 also code for an integrase domain protein but have targeted a protein-coding gene and are flanked by direct repeats (8 bp, CTGGCACA and 13 bp, GAACATGGTGTTC, respectively). This is reminiscent of the 10,007 bp insertion in strain NRC-1 compared to R1, which also codes for an integrase domain protein and shows an 8 bp direct repeat.
3.2.2 The patchy relationships between plasmids of strains 91-R6 and R1
The most prominent relationships between plasmids from strain R1 and strain 91-R6 are (a) 107 kb of pHSAL1 that are shared with pHS3 (Table 9), (b) 42.5 kb of pHS3 which match to part of divSEG12 from the 91-R6 chromosome (Tables 5 and 6), and (c) 13 kb that are related between pHSAL2 and R1 plasmids pHS1/pHS2 in their duplicated region (Table 8) (for details see below).
| Tag | Pos (91-R6; pHSAL2) | Length (91-R6) | GC (%) | Sequence comparison | Pos (R1; plasmid) | Length (R1) | Pos (NRC-1; plasmid) | Length (NRC-1) | Comment |
|---|---|---|---|---|---|---|---|---|---|
| p201 | 1–4,627 | 4,627 | 53.6 | – | – | – | – | – | Unique; MGE:multi |
| p202 | 4,628–8,483 | 3,856 | 54.7 | 96% |
pHS1 rev 8,806–12,661 |
3,856 |
pNRC100 rev 8,806–12,661 |
3,856 | dup(pNRC100 + 200) |
| p203 | 8,480–8,583 | 104 | 50.0 | 99% |
pHS1 fwd 58,308–58,411 |
104 |
pNRC100 fwd 57,459–57,562 |
104 | dup(pHS1 + 2); dup(pNRC100 + 200) |
| p204 | 8,584–20,422 | 11,839 | 56.8 | – | – | – | – | – | Unique; MGE:multi |
| p205 | 20,423–25,431 | 5,009 | 61.0 | 91% |
pHS1 rev 137,266–142,276 |
5,011 |
pNRC100 rev 123,875–128,885 |
5,011 | Dup(pHS1 + 4) |
| p206 | 25,432–33,747 | 8,316 | 59.1 | 99% (3 point mutations) |
pHS1 rev 126,522–137,265 |
10,744 |
pNRC100 rev 114,544–123,874 |
9,331 | Dup(pHS1 + 4); MGE:MITEHsal1 (R1:targeted by MGE:ISH8); targeted by MGE:ISH4 (R1, NRC-1) |
| 33,748–34,152 | 405 | – | – | – | – | – | – | MGE:MITEHsal2 + TSD | |
| p207 | 34,153–34,411 | 259 | 52.1 | 100% |
pHS1 rev 126,270–126,528 |
259 |
pNRC100 rev 114,292–114,550 |
259 | Dup(pHS1 + 4) |
| p208 | 34,412–63,436 | 29,025 | 56.6 | – | – | – | – | – | Unique; MGE:multi |
| p209 | 63,437–77,377 | 13,941 | 53.5 | Dup(pHSAL1 + 2); MGE:multi | |||||
| p210 | 77,378–79,715 | 2,338 | 63.7 | 83% |
pHS1 fwd 43,181–45,518 |
2,338 |
pNRC100 fwd 42,332–44,669 |
2,338 | Dup(pHSAL1 + 2); dup(pHS1 + 2); Dup(pNRC100 + 200) |
| p211 | 79,716–91,631 | 11,916 | 56.3 | – | – | – | – | – | Dup(pHSAL1 + 2) |
| 91,632–94,481 | 2,850 | 57.8 | – | – | – | – | – | Dup(pHSAL1 + 2); MGE:multi | |
| p212 | 94,482–94,543 | 62 | 54.8 | 100% |
pHS3 rev 247,793–247,854 |
62 | – | – | Dup(pHSAL1 + 2) |
| 94,544–95,942 | 1,399 | – | – | – | – | – | – | Dup(pHSAL1 + 2); MGE:ISHsal2 + TSD | |
| p213 | 95,943–99,055 | 3,113 | 58.4 | 100% |
pHS3 rev 244,109–247,797 |
3,689 | Dup(pHSAL1 + 2); targeted by MGE:ISH2 + TSD(R1;TSD 55 nt) | ||
| p214 | 99,056–101,544 | 2,489 | 50.7 | – | – | – | – | – | Dup(pHSAL1 + 2) |
| 101,545–102,666 | 1,122 | – | – | MGE:ISH1 + TSD (TSD only in pHSAL2) |
Note
- Plasmid pHSAL2 was split into subregions based on BLASTn results (excluding MGE sequences from BLASTn analysis). Junctions were chosen to reflect onset/offset of close matches to strain R1 (and NRC-1) and also to reflect the start of the 39 kb duplication between pHSAL1 and pHSAL2. Region tags start with p2 (plasmid pHSAL2) (tag). The comment column (column) term “unique” indicates the absence of a close homolog (>90% DNA sequence identity) in R1. From the onset of the 39 kb duplication between pHSAL1 and pHSAL2, this is indicated (“dup(pHSAL1 + 2)”). When the region carries MGEs, this is indicated (“MGE:multi”). For all matches, the plasmid is indicated (no matches to chromosomal sequences were encountered). It is also indicated if the match is to the forward (“fwd”) or reverse (“rev”) strand. If the homologous region is on a plasmid duplication in R1, the position is given for pHS1 and the duplication is indicated in the comment column (“dup(pHS1 + 2)” or “dup(pHS1 + 4)”). The duplicated region is also represented as “ppd” in Table 9. If the homologous region is on a plasmid duplication in NRC-1, the position is given for pNRC100 and the duplication is indicated in the comment column (“dup(pNRC100 + 200”). MGEs which are specific for strain 91-R6 are indicated, with TSDs indicated (“MGE:MITEHsal2 + TSD”).
- Special notes for certain regions: (a) Regions p202 and p203 overlap due to a shared tetranucleotide (TGGT). (b) Regions p205 and p206 are separate because region p206 is near-identical (99% DNA sequence identity, only three point mutations) while region p205 is highly homologous (91% DNA sequence identity). (c) Region p206 carries the MGE MITEHsal1 which is not targeted in strains 91-R6 and NRC-1 but is targeted in strain R1 by MGE:ISH8 (“MGE:MITEHsal1 (R1:targeted by MGE:ISH8”). This region is additionally targeted by a shared MGE:ISH4 in strains R1 and NRC-1 (“targeted by MGE:ISH4(R1,NRC-1)”). (d) Regions p208 and p209 are separate because region p209 marks the start of the pHSAL1/pHSAL2 duplication. (e) Region p213 is targeted in R1 by MGE:ISH2 which has an exceedingly long target duplication (55 bp) (“targeted by MGE:ISH2 + TSD(R1;TSD 55 nt)”). This is the element between p3G and p3H in Table 5. (f) At the 3′ end of the pHSAL1/pHSAL2 duplication is a MGE:ISH1. This MGE has a TSD in pHSAL2 but not in pHSAL1 (“MGE:ISH1 + TSD (TSD only in pHSAL2)”).
| Tag | Pos (91-R6; pHSAL1) | Length (91-R6) | GC (%) | Sequence comparison | Pos (R1; pHS3) | Length (R1) | Pos (NRC-1; pNRC200) | Length (NRC-1) | Comment |
|---|---|---|---|---|---|---|---|---|---|
| pp11 | 1–46,381 | 46,381 | 64.9 | Identical | 42,737–89,117 | 46,381 | 159,392–205,772 | 46,381 | Multiple MGEs |
| 46,382–46,755 | 374 | – | – | – | – | – | – | MITEHsal3 + TSD | |
| pp12 | 46,756–101,256 | 54,501 | 61.3 | One point mutation; one indel (4 nt) | 89,109–145,008 | 55,900 | 205,764–260,260 | 54,497 | R1: targeted by ISH3 |
| 101,257–102,885 | 1,629 | – | – | – | – | – | – | HsTy_ISNpe8 + TSD | |
| pp13 | 102,886–104,904 | 2,019 | 50.9 | Identical | 145,001–148,683 | 3,683 | 260,253–265,329 | 5,077 | R1 + NRC-1: targeted by ISH30; ISH30 additionally targeted by ISH3 in NRC-1 |
| 104,904–107,280 | 2,376 | – | – | – | – | – | – | ISH3D + TSD; targeted by ISH9 | |
| pp14 | 107,281–109,176 | 1,896 | 52.1 | Identical | 148,679–150,574 | 1,896 | 265,325–265,437 | 113 | NRC-1: targeted by ISH8 and truncated |
| ppd | 109,177–148,406 | 39,230 | 55.8 | – | – | – | – | – | Exact duplication between pHSAL1 and pHSAL2; multiple MGEs |
Note
- The unique region of plasmid pHSAL1 matches extensively to part of plasmid pHS3 from strain R1 and of plasmid pNRC200 from strain NRC-1. Region tags start with pp1 (plasmid/plasmid correlation for pHSAL1) (tag). The matching region covers four parts (pp11–pp14), which total to 107,860 bp. Except for targeting by MGEs and minor differences in region pp12, the sequences are identical. For strains R1 and NRC-1, sequence length includes targeting MGEs. The four parts are separated by strain 91-R6 specific MGEs, which are surrounding by target sequence duplications (“+TSD”). The remainder of pHSAL1 is the long duplication with pHSAL2 (ppd; d for duplication). This matches to “p209”–“p214” in Table 8. For the regions from pHSAL1, the GC content is given (“GC (%)”). High-GC regions are highlighted in green, low-GC regions in yellow.
The unique region of strain 91-R6 plasmid pHSAL1 corresponds to R1 plasmid pHS3
The unique region (107 kb) of plasmid pHSAL1 is exceedingly similar to part of pHS3 from strain R1. Most of this common sequence also occurs on pNRC200 from strain NRC-1 (Tables 9 and 5, tag pp11-pp14). The main differences are due to targeting by MGEs (three events in strain 91-R6, two in R1, and two in NRC-1). Leaving aside MGE targeting (and the deletion of 1.7 kb at the 3′ end in NRC-1), the common region covers 107,860 bp and is almost identical in all three strains, except that the type strain has one point mutation and one indel (four bases, intergenic). Such an extreme conservation is atypical for plasmids. The region is GC-rich (see Figure 1, Figure A1 in Appendix 2, and Table 9), shows a high protein coverage upon proteome analysis in strain R1 (Tebbe et al., 2005), and encodes several essential genes. Accordingly, the megaplasmids carrying this extended region can be considered minichromosomes (Pfeiffer, Schuster, et al., 2008). Among the genes which are encoded in this region, HBSAL_12005 to HBSAL_12615 sequence in strain 91-R6 is the only arginine–tRNA ligase (argS, HBSAL_12475) in each of the three strains. Adjacently encoded is the arginine fermentation cluster (arcDBCAR) which has been characterized in strain R1 (Ruepp & Soppa, 1996; Wimmer, Oberwinkler, Bisle, Tittor, & Oesterhelt, 2008) and is required for Halobacterium bioenergetics (Gonzalez et al., 2008). The chemotactic arginine sensor Car is encoded just beyond this shared region, in a 93.5 kb sequence which is unique to pHS3 (Table 5, tag p3F). The first step of arginine fermentation is the cleavage of arginine into ornithine and carbamoylphosphate. The latter compound is one of the substrates of the enzyme aspartate carbamoyltransferase (pyrBI), which catalyzes the first committed step of pyrimidine biosynthesis and is encoded immediately upstream of argS. This region also codes for other metabolically important enzymes, two of which have been characterized in Halobacterium: alkaline phosphatase, aph, (Wende et al., 2010) and catalase-peroxidase, katG (Long & Salin, 2001). Other important genes are glycerol dehydrogenase (gldA1), the queCED genes required for biosynthesis of the hypermethylated modified tRNA base archaeosine, and a probable siderophore biosynthesis cluster (iucABCD). Finally, this region codes for the so-called “chromosomal” gas vesicle cluster (gvpACDEFGHIJKLMNO) (Surek, Pillay, Rdest, Beyreuther, & Goebel, 1988). The assignment as “chromosomal” was a prediction based on the high GC content and was made before the genome structure had been resolved. The “plasmid” gas vesicle cluster is present in strains R1 and NRC-1 but not encoded in strain 91-R6 (see below). The last part of the 107 kb region shared between strains 91-R6 and R1 has been deleted from the plasmid in NRC-1 (see Table 9, tag pp14). However, upstream of this shared region, R1 and NRC-1 have a long region of 31 kb in common, which is lacking in strain 91-R6 (see Table 5, tag p3A). This region encodes the kdpFABCQ cluster for a potassium uptake system (see below). This region may have been lost in strain 91-R6 during the event which transferred the 3′ terminal part of the 39.2 kb duplication from pHSAL2 to pHSAL1 (Figure A2, junction JA2) (see Appendix 8).
A strain-specific chromosomal region from strain 91-R6 (divSEG12) shows a close but complex relationship to plasmid pHS3 from strain R1
A near-identical sequence of 42.5 kb (having only two point mutations) is shared between the 164 kb strain-specific chromosomal sequence divSEG12 and R1 plasmid pHS3. However, this sequence has become “scrambled” into four fragments by MGE targeting, genome inversions, and strain-specific deletions (Table 5, tag p3I, J, K, L; Table 6, tag c10,c11,c13,c16, Figure 2). The underlying evolutionary history and processes could be discerned by junction analysis, taking into account targeted or truncated genes as well as “hybrid TSDs” (target site duplications which became disjunct by a subsequent genome rearrangement). This analysis also uncovered a 3 kb extension of the shared sequence which, however, has been shifted from chromosomal divSEG12 to the duplicated part of pHSAL1/pHSAL2 (Table 5, tag p3G, H, see below). Full details are provided in Appendix 8 (junctions JC1 and JC2, see also Figures 3 and 4).
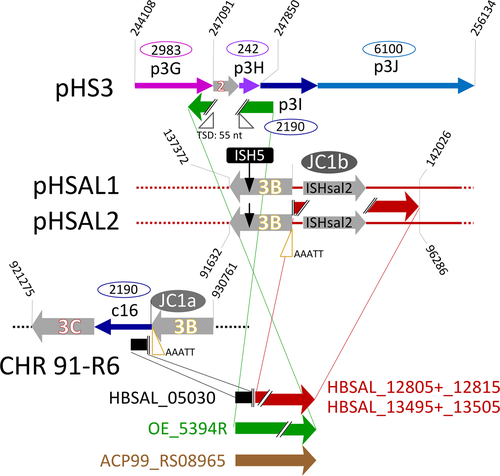
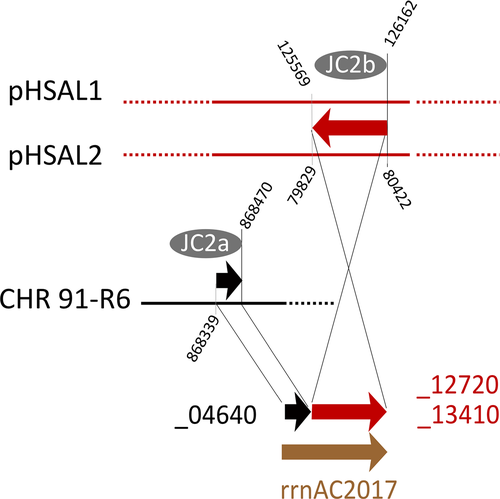
In Figure 2, the first two fragments of 2.1 kb and 6.1 kb are contiguous in pHS3 (Table 5, tag p3I, p3J) but dislocated and inverted in divSEG12 (Table 6, tag c10,c16) (Figure 2, junction JB1). Disconnection in divSEG12 is attributed to MGE targeting (ISH3C) with a subsequent MGE-triggered genome inversion. This attribution is supported by a hybrid TSD. PacBio reads reveal heterogeneity with respect to this inversion, both orientations being frequent in the population with support from at least 70 reads (see also below, Section 3.3, Figure 5, and Appendix 8 and Appendix 10, case D). (b) On divSEG12, the 6.1 kb fragment is contiguous with a 24.6 kb matching sequence (Table 6, tag c11) which is dislocated and inverted on pHS3 (Table 5, tag p3L; Figure 2, junction JB2, Figure 6). Disconnection and inversion in pHS3 is attributed to MGE targeting (ISH8B) with a subsequent MGE-triggered genome inversion. This attribution is supported by (i) two hybrid TSDs and (ii) one pair of disrupted proteins. OE_5405F and OE_5013R together correspond to HBSAL_04690 and are a full-length homolog of Halxa_0005. The N-terminal fragment, corresponding to OE_5405F, has been lost from strain NRC-1, while the C-terminal fragment, corresponding to OE_5013R, has been retained (VNG_6145a) (see below; for further details see Appendix 8). Finally, (iii) there is a disrupted transposon ISH32 where the two disconnected fragments together represent one complete MGE. (c) The 24.6 kb sequence p3L traverses the point of ring opening in pHS3. The ring opening point is associated with a discontiguity between R1 plasmid pHS3 and NRC-1 plasmids pNRC100 and pNRC200. While regions p3L and c11 are colinear between strains 91-R6 and R1, part of this sequence has been lost from NRC-1 (1,065 bp; including the equivalent to OE_5405F) and part has been shifted to pNRC100 (16,511 bp; reverse orientation). The region shared between divSEG12, pHS3, and pNRC100 codes for an arsenic resistance cluster which has been characterized (Wang, Kennedy, Fasiludeen, Rensing, & Dassarma, 2004). (d) The next and last common fragment has 6.3 kb (Table 5, tag p3K; Table 6, tag c13) but is still inverted on pHS3. In divSEG12, these fragments are separated by a 8.2 kb strain-specific sequence (Table 6, tag c12) with just one ISH2 element at the corresponding position in pHS3 (Figure 2, junction JB3). This is attributed to MGE targeting (integration of two copies of ISH2) with subsequent recombination of these MGEs, resulting in deletion of the intervening sequence. This attribution is supported by the absence of a TSD around the ISH2 and by completeness of HBSAL_04810 while its R1 equivalent OE_5019R is truncated at the ISH2 element and does not continue on the other side.
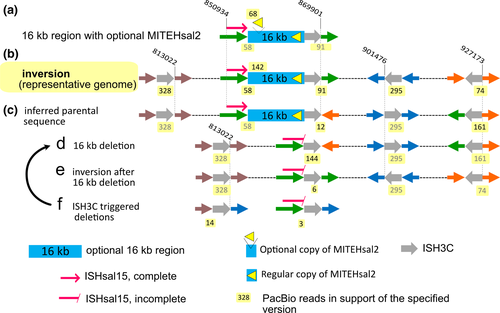
The sequence between the last common fragment and the dislocated and inverted first fragment on divSEG12 is separated by 12 kb (Table 6, tag c14 + c15 + MGE:ISH3). This region consists of a 6,038 bp sequence with 87% DNA sequence identity to part of the duplication between R1 plasmids pHS1 and pHS2. The adjacent 4,509 bp are specific for strain 91-R6, followed by the MGE of subtype ISH3C which is involved in the inversion.
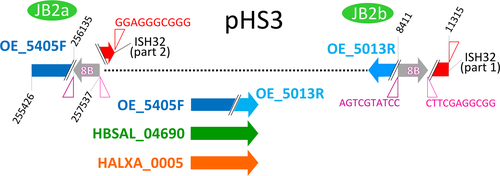
Targeted pseudogenes start in the strain-specific chromosomal region from strain 91-R6 (divSEG12) but continue in the region duplicated between pHSAL1 and pHSAL2
- The fragments of the first pseudogene, together, are a full-length homolog of WP_049986279.1 (ACP99_RS08965) from Halobellus rufus (these codes originate from NCBI) (Figure 3). In strain R1, this gene (OE_5394R) is encoded on pHS3 (region p3I + p3H + p3G, Table 5) and has been targeted by ISH2. Targeting resulted in a peculiar 55 bp target site duplication. In strain 91-R6, the gene (HBSAL_05030) has been targeted at a different position by transposon ISH3B (Figure 3, junction JC1; Appendix 8). While the N-terminal part remained on the chromosome, the C-terminal region has become part of the duplicated region of plasmids pHSAL1 and pHSAL2, again adjacent to an MGE of subtype ISH3B. On both plasmids, the C-terminal part (HBSAL_12805 + 12815; HBSAL_13495 + 13505) has been additionally targeted by a copy of transposon ISHsal2.
- The fragments of the second pseudogene, together, are a full-length homolog of rrnAC2017 from Haloarcula marismortui (Figure 4, junction JC2). The N-terminal part (HBSAL_04640) is encoded on the chromosome (Table 6, tag c09) and terminates only 42 nt from a transposon of type ISH3C upstream of region c10 in strain 91-R6 (Figure 2, Table 6; tag c10; Appendix 8). The C-terminal part (HBSAL_12720; HBSAL_13410) is encoded on the reverse strand of the duplicated part of plasmids pHSAL1 (gene start at nt 126,162) and pHSAL2 (gene start at nt 80,422) and is also not adjacent to a MGE. The chromosomal region c09, encoding HBSAL_04640, is part of the 16 kb sequence which has been deleted in part of the population (see below, Section 3.3, and Figure 5).
Strain 91-R6 plasmid pHSAL2 shows partial matches to R1 plasmid pHS1
The 102 kb plasmid pHSAL2 consists of a unique region (63.4 kb) and shares a 39.2 kb duplication with pHSAL1. Based on junction analysis, the 3′ end of the duplicated sequence belonged originally to pHSAL2 and has been transferred to pHSAL1 (see Appendix 8, junction JA2, and Figure A2 in Appendix 2). In the duplicated region is a 3.1 kb match (100% DNA sequence identity) to R1 plasmid pHS3 (see above). Even though duplications occur in the plasmids from all three analyzed Halobacterium strains, the duplications from the strain 91-R6 plasmids do not overlap with those from the R1/NRC-1 plasmids. At less than 90% DNA sequence identity, regions have to be considered independent. There is only one such homolog (2.3 kb, 83% identity) which occurs on duplicated regions of all three strains and codes for subunits of cytochrome bd ubiquinol oxidase (cydAB). The residual 28.2 kb of the duplicated as well as 45.4 kb of the pHSAL2 unique region are restricted to strain 91-R6. pHSAL2 has a 13.3 kb match to pHS1 (duplicated on pHS4; Table 8, p205 and p206) which consists of a 8.3 kb region with just three point mutations and one inserted MGE, while the remaining 5.0 kb have only 91% DNA sequence identity. This is reminiscent of a 30 kb duplication with sequence identity between R1 plasmids pHS1 and pHS4 and an adjacent 10 kb duplication which is much more dissimilar (98.5% DNA sequence identity). There is one additional 3.8 kb match (96% DNA sequence identity) between pHSAL2 and pHS1, but regionally disconnected in both strains.
3.2.3 Strain-specific proteins which are encoded on plasmids of strains 91-R6 and R1
To our knowledge, none of the proteins specific to the plasmids of strain 91-R6 has been implicated in any important biological process. Experimental evidence may reveal such examples, but to date, this strain has only rarely been studied. Also, no strain-specific proteins are assigned to pHSAL1 because most of it is not strain-specific (shared with R1 plasmid pHS3) and the remainder is duplicated on and thus can be assigned to pHSAL2. In strains R1 and NRC-1, however, plasmid-specific proteins with known and relevant function have been characterized (see below).
Strain-specific proteins from plasmids of strain R1
Strain-specific regions from plasmids of strain R1 (and NRC-1) code for several Orc paralogs and also contribute to the multiplicity of basic transcription factors (several paralogs of tfb and tpb genes). The “plasmid” gas vesicle cluster is specific to strains R1 and NRC-1. This cluster has been extensively characterized in strain NRC-1 (DasSarma, 1989, 1993; DasSarma, Damerval, Jones, & Tandeau, 1987; DasSarma et al., 1988, 2013; Halladay, Jones, Lin, Macdonald, & Dassarma, 1993; Halladay, Wai-Lap, & Dassarma, 1992; Jones et al., 1989; Jones, Young, & Dassarma, 1991; Tavlaridou, Faist, Weitzel, & Pfeifer, 2013; Tavlaridou, Winter, & Pfeifer, 2014; Winter, Born, & Pfeifer, 2018). Also, the kdp-type potassium-transporting ATPase (kdpABCF) is encoded on a plasmid region which is present only in R1 and NRC-1 (Kixmuller & Greie, 2012; Kixmuller, Strahl, Wende, & Greie, 2011; Strahl & Greie, 2008). Chemotactic sensing of arginine, which is mediated by the transducer encoded by car (Storch, Rudolph, & Oesterhelt, 1999), is exclusive to strain R1 as it is encoded on a region of pHS3 which is neither represented in strain 91-R6 nor in strain NRC-1.
3.2.4 Correlating the differences between the strain R1 and NRC-1 chromosomes to that of the type strain
Apart from differences related to ISH elements, the chromosomes of strains R1 and NRC-1 show only 12 differences: four point mutations, five single-base frameshifts, and three indels (Pfeiffer, Schuster, et al., 2008). In the present study, the sequence of strain 91-R6 was interrogated at the positions corresponding to frameshifts and indels (see Appendix 9). For all frameshifts in protein-coding genes, strain 91-R6 is consistent with R1. Strain NRC-1 has recently been resequenced as part of an experimental evolution study looking at genetic changes over 500 generations (Kunka et al., 2019), and the revised NRC-1 sequence is consistent with R1 at all frameshift differences (for details see Appendix 9).
Based on the 91-R6 sequence, it can be concluded that the 133 bp indel (divSEG02) is a deletion in R1, the 423 bp indel is a deletion in NRC-1, and the 10,007 bp indel is an insertion in NRC-1. This insertion has similar characteristics (encodes an integrase domain protein, targets a protein-coding gene, and is flanked by direct repeats) to those of divSEG14 and divSEG31 from strain 91-R6 (see above, Section 3.2).
3.3 Population heterogeneity and MGEs
3.3.1 Analysis of population heterogeneity
During the supervised genome assembly, we discovered that the PacBio long sequencing reads manifested significant population heterogeneity, including (a) the presence/absence of mobile genetic elements at certain genomic locations, (b) small-scale genome inversions triggered by MGEs, and (c) other MGE-triggered genome rearrangements as detailed in Appendix 10 (see also Figures 7-10, 5). This probably caused the failure of the nonsupervised genome assembly pipeline to close the replicons so that it terminated with 43 distinct contigs.
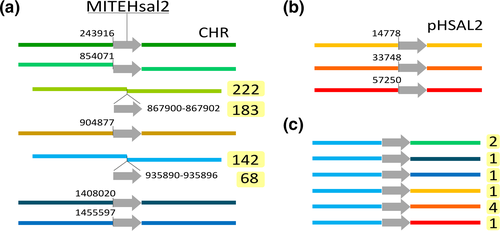
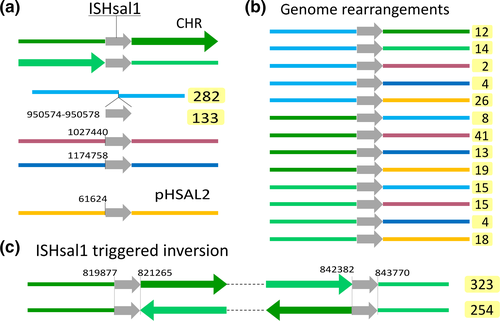
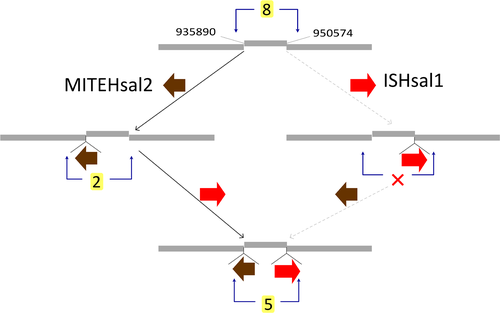
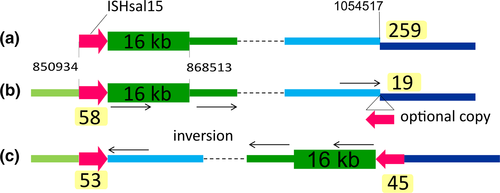
- a simple inversion of a 23.8 kb sequence positioned between oppositely oriented copies of the same transposon (ISHsal1, nt 819,877–843,770, including the two copies of the MGE; Figure 8; see Appendix 10 case B). Both orientations were supported by at least 250 PacBio reads, and the version selected was the one where the two fragments of a targeted pseudogene (HBSAL_04465 and HBSAL_04475) are adjacent to the same MGE and which retains a target site duplication (AGTTT) around one of these elements;
- two optional MGEs were detected, separated by a distance of only 14.6 kb. One was a copy of the transposon ISHsal1 (Figure 8) and the other (MITEHsal2) was a MITE (Figure 7). Genomic versions with zero or one MGE were supported by 133–282 PacBio reads, but only 15 PacBio reads traversed both MGEs. Of these, five contained both MGEs, and eight were devoid of both. Two reads contained only one of the MGEs, namely MITEHsal2, which suggests it integrated first (Figure 9). Other genome rearrangements involving these two MGEs occurred at low frequency (less than 20 reads, except for two cases) (Figures 7 and 8). Among these are cases where plasmid pHSAL2 has integrated into the chromosome.
- apart from a copy of ISHsal15 that occurs at nt 850,934–851,878, an optional additional copy was detected 202.6 kb away, integrated between nt 1,054,517 and 1,054,518. The majority of cases where the additional copy is present were associated with an inversion of the 202.6 kb intervening sequence (Figure 10).
- we consider it likely that the organism shifts its genome from an original form, to a slightly more streamlined genome version. This is associated with four closely spaced copies of ISH3C, where the 1st copy (nt 811,634–813,022, forward orientation) and the 3rd copy (nt 901,476–902,864, reverse orientation) are identical to each other and the 2nd copy (nt 868,513–869,901, forward orientation) and the 4th copy (nt 925,785–927,173, reverse orientation) are also identical to each other (Figure 5). Most of the region between the 2nd and the 4th copy (55.8 kb) corresponds to the 42.5 kb match between divSEG12 from strain 91-R6 and R1 plasmid pHS3 (see above, Section 3.2, Figure 2, and Appendix 8). For this 55.8 kb region, an inversion heterogeneity is observed in the population (Figures 2 and 5). The orientation selected for the representative genome (CP038631) disconnects divSEG regions c10 from c16 (as compared to pHS3 regions p3I and p3J), and thus it can be assumed that the inverted version is parental. At the right junction (4th copy), the parental sequence has a two-fold higher representation of PacBio reads (161) compared to the representative genome (74). At the left junction (2nd copy), however, the representative genome has strong support (91 reads) while the assumed parental sequence has only minimal support (only 12 reads; thus not suitable as a representative genome). The low number of reads at this junction can be attributed to a genome streamlining process which involves deletion of a 16 kb sequence. Many reads (144) support deletion of this 16 kb sequence, which thus seems to be gradually lost from the population.
The optional 16 kb sequence has several interesting features. (a) It carries the idiB gene, coding for isopentenyl-diphosphate delta-isomerase of type II. This gene seems dispensable as a type I isoform of this enzyme (idiA, HBSAL_09295) is encoded in the genome. (b) It carries (at its 3′ end) the disrupted gene HBSAL_04640, which represents only an N-terminal region. The C-terminal region is plasmid encoded (HBSAL_12720/HBSAL_13410) (see above, Section 3.2, Figure 4, and Appendix 8 junction JC2). (c) The 16 kb sequence carries a regular and also an optional copy of MITEHsal2 (Figure 5), which was a further complication for genome assembly. (d) The 16 kb sequence contains the only copy of transposon ISHsal16 so that its deletion cures the strain of this MGE. (e) At its left end, the 16 kb deletion extends into ISHsal15, thus truncating that element. This MGE has, however, escaped curing as there is an additional, optional copy, which is additionally involved in a 202 kb inversion (see under (c) and Figure 10).
The 55.8 kb invertible region covers the 3rd copy of ISH3C. While this is on the reverse strand in the representative genome, it is on the forward strand in the assumed parental sequence, thus forming a direct repeat with the upstream 1st and 2nd copies. As an additional population heterogeneity, deletions have been triggered by these direct repeats (see Figure 5). We encountered deletions involving the 3rd and 1st as well as involving the 3rd and 2nd copy (in its 16 kb deleted version) (Figure 5).
3.3.2 Mobile genetic elements in the three strains of Hbt. salinarum
A detailed analysis of the transposons and MITEs of the Hbt. salinarum type strain genome was performed (Table 7; see Appendix 11). This identified 17 novel types of transposons and 6 novel types of MITEs in strain 91-R6, all of which have been integrated into ISFinder (Siguier et al., 2012) (see Appendix 11). Overall, 15 MGEs are common, 10 occur only in the laboratory strains (R1 and NRC-1) and 21 occur only in the type strain (91-R6) (Table 7, Appendix 11). Strain-specific types of transposons and MITEs were likely introduced upon integration of foreign genetic material (plasmids or chromosomal genomic islands). This illustrates the high risk of being infected by MGEs when foreign genetic material is acquired by an organism.
4 DISCUSSION
This study has examined the genomic information carried by the type strain of the genus Halobacterium and explored its relationship to the two best studied strains of this species, R1 and NRC-1, both of which probably derive from the same isolate originally deposited at the culture collection of the National Research Council of Canada (NRC) (Grant et al., 2001; Pfeiffer, Schuster, et al., 2008). The comparative picture is that the strain 91-R6 chromosome shares a remarkably similar backbone with R1/NRC-1 (98.2%–98.8% ANIb, based on 1.85 Mb of shared sequence) but it differs significantly by several large replacements (genomic islands) as well as many smaller indels and replacements, and more than 6,700 point mutations. By contrast, the R1 and NRC-1 strains, which are laboratory variants derived from one original isolate, have chromosomes with only 12 differences (Pfeiffer, Schuster, et al., 2008) besides those associated with MGEs. The high in silico DDH values between the strains (95%) is well above the taxonomic threshold for membership of the same species (70%). For comparison, the two sequenced strains of Haloquadratum walsbyi (C23T and HBQS001), isolated in Australia and Spain, respectively, have an in silico DDH of 84.2%. While the results confirm that strain 91-R6 is an independent isolate from strains R1/NRC-1, their close similarity raises new questions. Does the conserved backbone indicate a species that (a) is particularly slowly evolving, (b) has high geographical mobility so that dominant strains rapidly spread and outcompete the microbial flora of distant hypersaline niches (regionally/globally), or (c) reflects a common source for both isolates. Regarding the relative rate of divergence, it may be significant that the NCBI taxonomy lists only six species of Halobacterium (https://www.ncbi.nlm.nih.gov/Taxonomy) despite its ease of cultivation and decades of isolation studies, while more recently discovered genera with similar cultivability have far more described species (Halorubrum, 46; Haloferax, 19; Haloarcula, 16), with new species reported frequently (http://www.bacterio.net/halorubrum.html). Two of the six listed Halobacterium species may not even represent contemporary examples as they were recovered from ancient rock salt (noricense, hubeiense). Regarding the “common source” hypothesis, strain 91-R6 was a Canadian isolate recovered from salt used for tanning of hides, and the parent strain of R1/NRC-1 was also an early member of the Canadian culture collection (NRC), and so could have originated from a similar source. Unfortunately, with the closure of the NRC culture collection, the records for NRC-1 were lost (Grant et al., 2001). More extensive genomic surveys of this species from the existing isolates in culture collections and from new isolates around the world should resolve this issue. For example, the type strain of “Hbt. cutirubrum” (now Hbt. salinarum), strain 63-R2 (NRC 34001, ATCC 33170, DSM 669), was isolated from salted buffalo hides by Lochead at the same time as strain 91-R6, and could provide further insights into strain diversity.
Much of the early work on strains of this species focused on the extraordinarily high mutation rates of genes for visible phenotypes, such as cell color and gas vesicle synthesis (DasSarma et al., 1988; DasSarma, Rajbhandary, & Khorana, 1983). Changes in these genes were found to be driven by transposons (insertion elements), with mutant frequencies as high as 1% (DasSarma, 1989; Jones et al., 1989), and suggested a rapidly evolving species. Transposition bursts could also be triggered by environmental stress (Pfeifer & Blaseio, 1990). However, later work determined the average genomic mutation rate of Hbt. salinarum NRC-1 to be very low, with 1.67 × 10−3 mutations per genome per replication (Busch & DiRuggiero, 2010), indicative of a high-fidelity replicative system. The disparity between MGE-related and MGE-independent mutation rates is a curious phenomenon, but the high polyploidy of haloarchaea and their rapid rates of gene conversion (Soppa, 2011; Zerulla & Soppa, 2014), coupled with multiple modes of gene exchange (Abdul Halim, 2013; Demaere et al., 2013; Erdmann, Tschitschko, Zhong, Raftery, & Cavicchioli, 2017; Rosenshine & Mevarech, 1991) may act to maintain genomic stability despite the high activity of MGEs. The dominance of MGE-related over MGE-independent mutations was also seen in a recent 500 generation experimental evolution experiment (Kunka et al., 2019). In the current study, a culture of the type strain was directly analyzed by long-read sequencing without prior colony purification, allowing observation of the types of variants that naturally accumulate. Numerous variants were detected, all of which could be ascribed to MGEs. These included simple MGE insertions, deletions (up to 16 kb), inversions (up to 202 kb), and other rearrangements. This pattern is consistent with the early studies of mutation in Hbt. salinarum, and with genome differences reported for strains R1 and NRC-1, the majority of which were found to be ISH-related (Pfeiffer, Schuster, et al., 2008). The variant reads for strain 91-R6 also revealed mergers between plasmid and chromosomal sequences, which is possible because haloarchaeal chromosomes often carry multiple, active replication origins, and additional ori sequences from plasmid integration are not disruptive. For example, the large plasmid pHV4 of Hfx. volcanii can stably insert into the main chromosome (Ausiannikava et al., 2018; Hawkins, Malla, Blythe, Nieduszynski, & Allers, 2013) taking with it an origin of replication. Our analyses can, however, not distinguish between different genome variants in distinct cells or variants within a single cell, although the latter is less likely to be encountered. A remarkable observation regarding plasmid pHSAL1 of strain 91-R6 is the extreme conservation to plasmid pHS3 of strain R1, with only two point mutations in 107 kb of shared sequence. On the other hand, part of pHS3 is found on a strain-specific chromosomal sequence in the strain 91-R6, again suggesting plasmid insertion into chromosomes is a common occurrence.
Three major genomic islands were detected, GI-1, 2, and 3 (corresponding to divSEGs 04, 12, 18), which together totaled 289 kb. They were characterized by lower than average %G + C, and increased densities of transposons and mCTAG motifs, although these differences were less intense in GI-3. The CTAG tetranucleotide is strongly avoided outside GIs but the physiological role of this modification is yet unresolved. In Haloferax, CTAG methylation has been ascribed to the ZIM methyltransferase which is conserved in haloarchaea (Ouellette et al., 2018).
GI-1 and GI-2 carry replication genes (Orc paralogs) and GI-2 has genes encoding ParA (partition) and toxin-antidote proteins (maintenance), suggesting a plasmid origin for both these regions. GI-1 replaces the well-known AT-rich island in the R1 and NRC-1 genomes while GI-2 is the longest strain-specific chromosomal sequence (164 kb), and at its termini it contains a split homolog of the strain R1 dmsA gene. A considerable part of GI-2 (42.5 kb) is virtually identical to R1 plasmid pHS3, with both sequences having suffered genome rearrangements. GI-1 and GI-2 also carry genes for secreted glycoproteins, glycosylation or both, while GI-2 possesses genes for defence against foreign DNA (BREX, restriction-modification). GI-3 (divSEG18) is a replacement and is rather different in nature to GI-1 and -2. It carries various biologically important or even essential genes which thus exist as distant homologs in the type and the laboratory strains. GI-3 includes many genes involved in protein N-glycosylation, which together with functionally related genes found in GI-1 and -2, could provide an altered glycan structure of the S-layer glycoprotein and other surface structures, possibly to evade virus predation.
Our analyses revealed that HBSAL_01455, a PilA-like protein of strain 91-R6 with very high similarity to the conserved type III signal sequence of Haloferax PilA proteins, is a regular gene in the biofilm forming strains 91-R6 and R1 (OE_1186A1F), while the corresponding gene has been targeted by a transposon and thus is disrupted in NRC-1, a strain that is not able to form biofilms under the conditions tested (Losensky et al., 2017, 2015).
The laboratory strains of Halobacterium lack a CRISPR-Cas defence system (as do about half of the haloarchaea with completely sequenced genomes). Absence of a CRISPR-Cas system has also been confirmed for the type strain 91-R6. However, the recently identified BREX virus defense system (subtype 5) was identified on plasmid pHS3 of strain R1, in a region which is absent from strain NRC-1. One gene attributed to type 5 BREX systems is a helicase domain protein named BrxHII, which is disrupted in strain R1, making it uncertain if the system is functional. While the BREX genes are not highly conserved in strain 91-R6, this strain codes for a distantly related BREX system on genomic island GI-2. A helicase domain protein BrxHII could not be identified, and methylation of A residues was not detected in PacBio reads, so that the functionality of this system is also uncertain.
A remarkable feature of strain 91-R6 is that it carries a large set of strain-specific MGEs (transposons and MITEs), even though only a relatively small part of the genome is unique when compared with strains R1 and NRC-1. Most of the novel MGEs are found on the GIs and in the strain-specific plasmid pHSAL2. This is consistent with the notion that the acquisition of foreign genetic material is likely to bring novel MGEs that can infect other sites of the genome.
Finally, the sequencing of the type strain can be seen as contributing to projects such as the Genomic Encyclopedia of Bacteria and Archaea (GEBA) and its follow-up projects (see www.dsmz.de/research/bioinformatics/phylogenomics/projects) that aim to systematically sequence the archaeal and bacterial branches of the tree of life (Wu et al., 2009). Our efforts not only provide the genome sequence of the “type species of the type genus of the family and the order” (Oren, 2012), but also a high-quality reference annotation and comprehensive comparison to closely related strains, which are expected to be useful and relevant resources for the scientific community.
ACKNOWLEDGMENTS
We thank Elisabeth Bruckbauer for help with growing the strain and PCR analyses. This research received no specific grant from any funding agency in the public, commercial, or non-for-profit sectors.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
A.M. and F.P. conceptualized the study; F.P. performed data curation; F.P. was the project administrator; F.P. and M.D-S. performed formal analysis; B.H. was involved in funding acquisition; A.M., F.P., M.D-S., and G.L. contributed to investigation and validation; A.M. provided the resources; B.H. supervised the entire proceedings; F.P., M.D-S., and G.L. contributed to visualization; F.P. and M.D-S. were involved in writing the original draft; A.M., B.H., F.P., M.D-S., and G.L. contributed to writing, reviewing and editing of the final draft.
ETHICAL APPROVAL
None required.
APPENDIX 1
| Species |
Hs 91-R6 [CP038631] |
Hs R1 [AM774415] |
Hs NRC-1 [AE004437] |
Hbt. jilantaiense |
Hbt. sp. DL1 [CP007060] |
Hbt. hubeiense [LN831302] |
Hbt. noricense CBA1132 |
|---|---|---|---|---|---|---|---|
| Hs 91-R6 | |||||||
| Hs R1 | 95.2 | ||||||
| Hs NRC-1 | 95.1 | 99.7 | |||||
| Hbt. jilantaiense | 24.6 | 24.1 | 24.1 | ||||
| Hbt. sp. DL1 | 21.4 | 21.4 | 21.4 | 22.4 | |||
| Hbt. hubeiense | 21.2 | 21.2 | 21.2 | 22.9 | 23.9 | ||
|
Hbt. noricense CBA1132 |
20.9 | 20.8 | 20.8 | 22.2 | 23.4 | 37.2 |
Note
- The table contains in silico DNA–DNA hybridization (DDH) values calculated with the Genome-to-Genome Distance Calculator (GGDC) 2.1 at http://ggdc.dsmz.de/ggdc.php#. As recommended, formula 2 values are shown. The chromosomes of the three analyzed strains of Hbt. salinarum and, in addition, from other species from the genus Halobacterium are included. In the top row, the sequence accessions are given in square brackets. Yellow highlighting shows values above the species cutoff (70%).
| Hs 91-R6 | Hs R1 | Hs NRC-1 | Hbt. jilantaiense | Hbt. sp. DL1 | Hbt. hubeiense |
Hbt. noricense CBA1132 |
|
|---|---|---|---|---|---|---|---|
| Hs 91-R6 | 98.83 [88.87] | 98.84 [88.08] | 80.92 [63.53] | 77.18 [53.16] | 77.59 [57.31] | 76.95 [53.87] | |
| Hs R1 | 98.22 [74.83] | 99.62 [91.09] | 80.28 [52.17] | 77.08 [46.57] | 77.69 [50.55] | 76.47 [45.63] | |
| Hs NRC-1 | 98.23 [78.10] | 99.99 [97.21] | 80.29 [53.62] | 77.25 [47.63] | 78.02 [51.26] | 76.63 [46.86] | |
| Hbt. jilantaiense | 80.59 [50.84] | 80.19 [50.71] | 80.25 [50.46] | 78.64 [55.38] | 79.15 [53.39] | 78.62 [52.52] | |
| Hbt. sp. DL1 | 76.62 [39.48] | 76.48 [40.38] | 76.53 [39.53] | 78.34 [50.22] | 79.52 [48.81] | 79.26 [46.89] | |
| Hbt. hubeiense | 77.27 [40.82] | 77.34 [42.77] | 77.37 [41.70] | 79.22 [47.13] | 80.14 [46.47] | 88.13 [62.58] | |
|
Hbt. noricense CBA1132 |
76.81 [46.28] | 76.62 [46.96] | 76.68 [46.63] | 78.79 [56.41] | 79.77 [54.51] | 88.61 [75.14] |
Note
- Values in square brackets are the percentage of aligned nucleotides between the two chromosome sequences. The chromosomes of the three analyzed strains of Hbt. salinarum and, in addition, from other species from the genus Halobacterium are included. Light yellow highlighting shows values above the species cutoff (95%). Dark yellow highlighting is used for values above 80%. For accessions see Table A1.
- ANIb (average nucleotide identity, BLASTn) values calculated at http://jspecies.ribohost.com/jspeciesws
| Strain | Replicon | Length | Protein-coding genes | Spurious ORFs | Locus tag series | First; last serial number |
|---|---|---|---|---|---|---|
| R1 | Chromosome | 2,000,962 | 2,151 | 5,335 | OE_1 to OE_4 | 1001;4759 |
| R1 | pHS1 | 147,625 | 168 | 227 | OE_7 | 7001;7224 |
| R1 | pHS2 | 194,963 | 220 | 366 | OE_6 | 6001;6358 |
| R1 | pHS3 | 284,332 | 291 | 524 | OE_5 | 5001;5448 |
| R1 | pHS4 | 40,894 | 38 | 68 | OE_8 | 8001;8050 |
| R1 | total | 2,668,776 | 2,868 | 6,520 | ||
| NRC-1 | Chromosome | 2,014,239 | 2,174 | 26 | VNG_0 to VNG_2 | 0001;2679 |
| NRC-1 | pNRC100 | 191,346 | 223 | 20 | (VNG_7) | 7001;7176 |
| NRC-1 | pNRC200 | 365,425 | 420 | 18 | VNG_6 | 6001;6487 |
| NRC-1 | total | 2,571,010 | 2,817 | 64 | ||
| 91-R6 | Chromosome | 2,178,608 | 2,346 | 0 | HBSAL_00 to HBSAL_11 | 00005;11730 |
| 91-R6 | pHSAL1 | 148,406 | 170 | 0 | HBSAL_12 | 12005;12850 |
| 91-R6 | pHSAL2 | 102,666 | 108 | 0 | HBSAL_13 | 13005;13540 |
| 91-R6 | Total | 2,429,680 | 2,624 | 0 |
Note
- For the classification of ORFs as being protein-coding or spurious see Appendix 4 and (Pfeiffer, Schuster, et al., 2008). Spurious ORFs in strain R1 are hidden internal records, used for annotation surveys (including proteomic analyses and ORF correlation between R1 and NRC-1), which are skipped upon genome submission. The term “locus tag series” refers to a set of serial numbers in the “thousands” indicated by the digit, with first and last number of the series specified in the adjacent column (e.g. ORFs from R1 plasmid pHS2 have locus tags from OE_6001R to OE_6358F). The are no spurious ORFs in strain 91-R6 because gene calling had been extensively surveyed, including removal of spurious ORFs, prior to locus tag assignment.
| Original code | Corrected code | Comment |
|---|---|---|
| VNG_0240a | VNG_0243a | |
| VNG_0287a | ||
| VNG_0335a | N-term part of a targeted gene | |
| VNG_0335b | VNG_0337a | C-term part of a targeted gene |
| VNG_0475a | ||
| VNG_0772a | VNG_0771a | |
| VNG_0892a | ||
| VNG_0950a | ||
| VNG_1173a | ||
| VNG_1390a | ||
| VNG_1534a | ||
| VNG_1598a | ||
| VNG_1675a | ||
| VNG_1818a | ||
| VNG_1886a | ||
| VNG_1964a | VNG_1963a | |
| VNG_2081a | ||
| VNG_2298a | ||
| VNG_2466a | ||
| VNG_2608a | ||
| VNG_2644a |
Note
- These locus tags were encountered when the genomes of strains NRC-1 and R1 had been compared (Pfeiffer, Schuster, et al., 2008). As described in Appendix 5, some of these locus tags had a serial number which deviated from that of the preceding ORF, as detected by script-based checking. The locus tags were replaced so that the serial number is taken from the preceding ORF.
APPENDIX 2
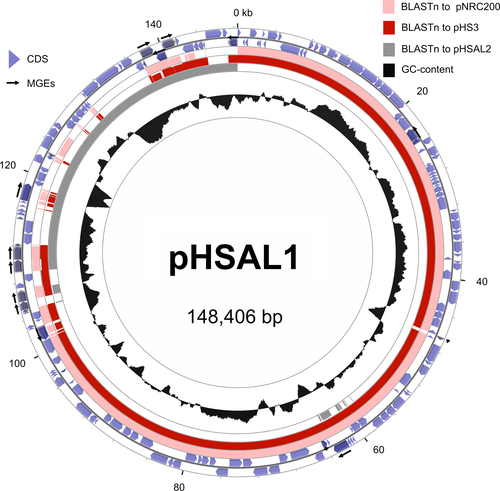
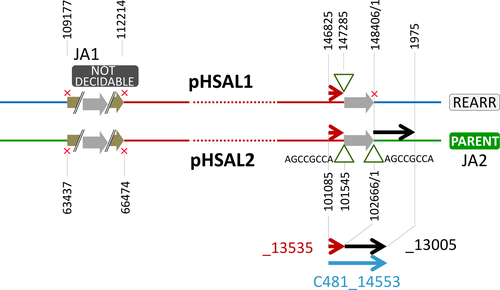
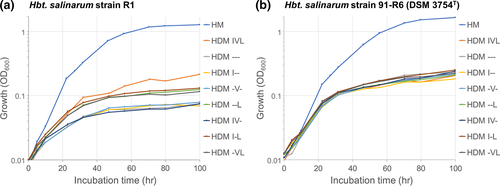
APPENDIX 3
REPLICONS (CHROMOSOMES AND PLASMIDS) OF THE THREE ANALYZED HALOBACTERIUM STRAINS (91-R6, R1, AND NRC-1)
This text lists the replicons (chromosome and plasmids) of the three strains (91-R6, R1, and NRC-1) at the DNA level. The reference accessions for these replicons are CP038631.1, CP038632.1, and CP038633.1. For protein-coding genes and other annotation issues see below (Appendix 4, Appendix 5, and Appendix 6).
All three strains of Hbt. salinarum have one major chromosome with a high GC content and 2–4 large plasmids (or minichromosomes) with a diminished GC content (Tables 2 and 3). The chromosomes are between 2.0 and 2.2 Mb in length and have a GC content of 67%–68%. The plasmids are between 40 and 365 kb and have a GC content of 56%–60% except for pHSAL1 (60.6%).
The chromosomes are highly similar to each other and completely colinear with a small set of strain-specific sequences (Table 4). Roughly half of these are strain-specific copies of mobile genetic elements (MGEs). Other strain-specific sequences may be up to 164 kb in length and several of the longer ones have characteristics which are typical for plasmids (e.g. diminished GC content, many MGEs). The chromosomes of strains R1 and NRC-1 are exceedingly similar with only 12 differences (four point mutations, five single-base frameshifts, and three indels of 133 bp, 423 bp, and 10,007 bp) (Pfeiffer, Schuster, et al., 2008). All other differences reflect strain-specific transposon targeting or point mutations within transposons.
All plasmids show extensive interplasmid duplications ranging from 30 to 112 kb. Plasmids pHSAL1 and pHSAL2 from strain 91-R6 share a 39,230 bp duplication (pHSAL1: 109,177–148,406; pHSAL2: 63,437–102,666; both regions mark the 3′ end of the plasmid). The plasmid duplications in strain R1 have been previously reported (see Table S2 of (Pfeiffer, Schuster, et al., 2008)). They are: (a) a 61,818 bp perfect duplication between pHS1 and pHS2 (pHS1:37,110–98,927; pHS2: 86,549–148,366); (b) a 30,099 bp perfect duplication between pHS1 and pHS4 (pHS1:113,272–143,370; pHS4:9,651–39,749); (c) an imperfect 9,740/7,316 bp duplication with 98.5% DNA sequence identity between pHS1 and pHS4 (pHS1:103,532–113,271; pHS4:2,335–9,650).
The duplications between plasmids pNRC100 and pNRC200 of strain NRC-1 (taken from Table S2 of (Pfeiffer, Schuster, et al., 2008) are: (a) 112,795 bp perfect duplication (pNRC100:1–112,795; pNRC200:1–112,795); (b) a near-perfect inverted 32,633 bp duplication within pNRC200 (pNRC200: 32,043–64,675/forward strand; pNRC200:365,424–332,793/reverse strand). The sequences differ by a one-base frameshift, which disrupts the start codon in an ISH3-type transposase in the inverted copy of the repeat. (c) a longer version of the near-perfect inverted duplication (39,168 bp) within pNRC100 (pNRC100:32,043–71,210/forward strand; pNRC100:191,345–150,254/reverse strand). In the extended region, this duplication has an extra copy of a transposon. In the sequence shared with the inverted duplication on pNRC200, this sequence has the same one-base frameshift which disrupts a transposase gene. Thus, a given sequence may occur four times in the plasmids of strain NRC-1 (in the longer version of the inverted repeat), three times (in the shorter version of the inverted repeat) or two times (in the duplication outside the inverted repeat).
Due to a partial overlap of the plasmid duplications in strains R1 and NRC-1, the number of copies varies from 2 (one copy in each strain) to 6 (two copies in R1, four copies in NRC-1). Both possibilities leading to five copies have been encountered: four copies in NRC-1 but only one in R1 or two copies in R1 and three in NRC-1.
Despite major differences in the overall structure of the plasmids from strains R1 and NRC-1, they share 350 kb of unique common sequence with only few sequence differences (except for strain-specific transposon targeting) (Pfeiffer, Schuster, et al., 2008). However, there are strain-specific plasmid sequences. While such strain-specific sequences are substantial in R1 plasmids (totaling to 210 kb), they are only minor in NRC-1 plasmids (5 kb).
Only a minor region of the duplication in strain 91-R6 plasmids pHSAL1 and pHSAL2 matches to a plasmid from strain R1, and this region is duplicated neither in the R1 nor in the NRC-1 plasmids.
APPENDIX 4
BASIC PRINCIPLES OF ORF CALLING: GENES, PSEUDOGENES, AND SPURIOUS ORFS
Protein-coding genes, gene calling, and start codon assignment
Protein-coding genes correspond to open reading frames (ORFs) in the genome sequence. Various gene callers are available with varying performance on GC-rich genomes. For strain 91-R6, gene calling was performed by GenMarkS-2 (Lomsadze et al., 2018), an up-to-date gene caller with good performance for GC-rich genomes. However, initial gene calls were subjected to extensive curation based on principles developed for the genome from strain R1 (Pfeiffer & Oesterhelt, 2015; Pfeiffer, Schuster, et al., 2008). For a considerable subset of the protein-coding genes of strain R1, start codon assignments could be based on proteomic data that was directed, in part, to identification of N-termini (Aivaliotis et al., 2007; Falb et al., 2006; Klein et al., 2007, 2005; Tebbe et al., 2009). The other major tool is homology-based analysis (Pfeiffer, Broicher, et al., 2008; Pfeiffer & Oesterhelt, 2015) which is supported by a dense occupancy of the sequence space, with >100 haloarchaeal genome sequences that are now available. To overcome missing gene calls, “intergenic” regions were compared to a large-scale protein sequence database (NCBI:nr) using BLASTx. A major effort was invested in the present project to ensure consistency of protein-coding gene annotation, including start codon assignment, between the three strains of Hbt. salinarum.
ORF classification as protein-coding gene or spurious ORF
In addition to protein-coding genes, additional frames may fortuitously remain open in the genome. We refer to such fortuitous open frames as “spurious ORFs” (Aivaliotis et al., 2007; Pfeiffer, Broicher, et al., 2008; Pfeiffer & Oesterhelt, 2015; Pfeiffer, Schuster, et al., 2008). Because spurious ORFs are especially prominent in GC-rich genomes, a considerable number of these were encountered in Halobacterium and had to be resolved upon cross-strain mapping of protein-coding genes and other ORFs.
Disrupted genes (pseudogenes, nonfunctional genes)
In nearly all prokaryotic genomes, some genes are found to be disrupted. Such genes carry a “pseudo” flag in the GenBank annotation. We prefer to call them disrupted genes because a small set of typical biological events leads to gene disruption, and many of these leave all transcription and translation signals intact. It thus can be assumed that many disrupted genes are transcribed and even translated, leading to aberrantly expressed forms of the protein. Their fate depends on the severity of the disruption. In extreme cases, the “disrupted” gene may code for a stable or even functional protein. In GenBank, disrupted genes are not translated and thus are not represented in the UniProt protein sequence database. In HaloLex, we attempt to associate a disrupted gene with a protein sequence which most closely reflects that of the assumed functional parent (Pfeiffer, Broicher, et al., 2008).
Typical biological sources of gene disruption are (a) in-frame stop codons, (b) frameshifts, (c) targeting by transposons or other MGEs, (d) terminal deletions resulting in ORF remnants that lack start or stop codons, or both, and (e) internal deletions, so that only terminal sequences are retained. It should be noted that cases (a), (b), and (c) lead to noncontiguous ORFs. While these are annotated as a single multiregion ORF in HaloLex, MGE targeting may result in the annotation of multiple independent ORFs in other annotation systems (see (Pfeiffer & Oesterhelt, 2015) for a discussion of this subject). In such cases, ORF correlation becomes complex. Thus, the correlation of nonfunctional ORFs required special efforts.
In the case of transposon targeting, the gene is split into two (or more) noncontiguous fragments. Annotation xmlstyle differs between strains R1 on one hand and strains 91-R6 and NRC-1 on the other hand. For R1, targeted genes are annotated as a single CDS which consists of multiple regions. For strains 91-R6 and NRC-1, the N-terminal and C-terminal fragments are annotated as distinct CDS, each consisting of a single region. Even though the multiregion representation is considered to reflect the biology more correctly, this has proven to cause major problems upon interaction with nucleic acid sequence databases (EMBL/GenBank) and thus was not adopted for the other two strains.
APPENDIX 5
ANNOTATION SOURCES AND ORF LOCUS TAGS FOR THE THREE ANALYZED HALOBACTERIUM STRAINS (91-R6, R1, AND NRC-1)
An overview about the replicons and the associated locus tags for the three Halobacterium strains is shown in Tables 2, 3 and Table A3 in Appendix 1.
General principles for the concerted annotation of protein-coding genes
The genomes of the three Halobacterium strains (especially their chromosomes) are exceedingly similar at the DNA sequence level. The majority of the sequences show 100.0% DNA sequence identity between strains R1 and NRC-1 and >99% DNA sequence identity between strains R1 and 91-R6. As a general rule for matching sequences, every protein-coding gene annotated in one strain must correspond to a partner gene in the other strain. Also, this gene pair must have a consistent start codon assignment (see also Appendix 4). The same principles apply to large-scale duplicated plasmid regions within the same strain.
Annotation source and locus tags for strain R1
For strain R1, proteins (ORFs) were extracted from Halolex (Sep 2018) and represent an up-to-date annotation based on our Gold Standard Protein based strategy (Pfeiffer, Broicher, et al., 2008; Pfeiffer & Oesterhelt, 2015; Pfeiffer, Schuster, et al., 2008). Proteins are encoded on the main chromosome and on four plasmids. Locus tags have the prefix OE, followed by an underscore, a serial number, and an extension. While the underscore between OE and the serial number was not used in the initial publication, it was recently added during an EMBL annotation update for consistency with current standards for ordered locus tags. Each extension may consist of or ends with a letter indicating the coding strand (F: forward; R: reverse). Between the serial number and the strand indicator may be a letter-integer combination to allow intercalation of postpredicted ORFs. Serial numbers above 5,000 indicate plasmid-encoded ORFs (for the correlation of replicon and locus tag series see Table A3 in Appendix 1).
ORF calling and curation of protein-coding genes from strain 91-R6
Initial gene prediction was performed by GenMarkS-2, an ORF caller which copes well with GC-rich genomes (Lomsadze et al., 2018). ORFs were exported in GenBank format with temporary ORF tags assigned. A detailed mapping of these ORFs to those from strain R1 and vice versa was performed. As outlined above, the two genome annotations were curated in parallel in order to ensure complete annotation consistency. For sequences which are specific for strain 91-R6, the GenMarkS-2 ORFs were subjected to curation according to our annotation principles (Pfeiffer & Oesterhelt, 2015). When gene disruption was encountered, this was resolved by taking appropriate measures. After completion of this effort, we performed an additional systematic attempt to identify and resolve residual missing gene calls. For this purpose, all intergenic regions ≥50 bp were subjected to BLASTx analysis against the NCBI:nr database. Identified protein-coding genes, eventually disrupted, were added to the annotation. Up to this point, only temporary locus tags had been in use.
Once this extensive curation effort had been completed, all protein-coding genes were assigned a locus tag. Because locus tags were only assigned after having missing gene calls resolved, no serial number intercalations were required. Locus tags with serial number above 12,000 are from the plasmids of strain 91-R6.
All protein-coding genes from strain 91-R6 are either directly correlated to those from strain R1 (Tables S1 and S2 (via Zenodo; https://doi.org/10.5281/zenodo.3528126)), or are strain-specific (Tables S3 and S5 (via Zenodo)) but may be homologous at reduced sequence similarity to proteins from strain R1. Strain-specific protein-coding genes from strain R1 are also listed (Tables S4 and S6 (via Zenodo)).
Annotation source and locus tag assignments for strain NRC-1
For strain NRC-1, the sequence of plasmid pNRC100 was published first (Ng et al., 1998), followed 2 years later by the main chromosome and plasmid pNRC200 (Ng et al., 2000). The genome sequence was downloaded from GenBank (Sep 2018) (chromosome: AE004437; pNRC200: AE004438; pNRC100: AF016485). The annotations appeared to reflect those originally submitted, without any subsequent annotation updates. For ORFs on the main chromosome and on plasmid pNRC200, locus tags with a VNG prefix are assigned. The underscore separator between VNG and the serial number was not used in the initial publication but was recently added by NCBI for consistency with current standards for ordered locus tags. Serial numbers above 6,000 indicate ORFs encoded on pNRC200 (Ng et al., 2000). For plasmid pNRC100, ORF numbers with prefix H were used in the original publication but locus tags of the VNG type were assigned neither in the original publication nor in the subsequent publication of the complete genome (Ng et al., 1998, 2000). Such locus tags had been assigned by NCBI in an early version of NC_001869, using serial numbers above 7,000. However, these have disappeared because NC_001869 was revised to have VNG_RS serial numbers (_RS locus tags are nowadays standard in RefSeq). Some of the VNG_7 series locus tags were retained in NC_001869 in the “old_locus_tag” field. We initially assigned simple serial numbers (from 1 to 176) for pNRC100 ORFs annotated in AF016485. Subsequently, these were replaced by VNG_7-type locus tags if the ORF could be positionally correlated with a RefSeq ORF that had an associated old_locus_tag. This resulted in the assignment of VNG_7 series locus tags for 132 of the 176 ORFs. In all cases, our simple serial number was identical to the last three digits of the VNG_7 series locus tag found in RefSeq. This analysis thus uncovered the VNG_7 assignment rules for the early version of NC_001869. We applied this rule to the residual 44 ORFs. It should be noted that UniProt has independently assigned locus tags for proteins encoded on pNRC100. They opted for the VNG_5 series, but these have not been further considered in our efforts. There were additional RefSeq modifications which were only temporarily available in the early RefSeq version of the NRC-1 chromosome (NC_002607): some missing gene calls had been resolved and locus tags with an “a” extension had been assigned by NCBI. From our previous analyses (Pfeiffer, Schuster, et al., 2008), we were aware of 20 such a-type locus tags and these were initially retained upon genome re-annotation (Table A4 in Appendix 1). When resolving additional missing gene calls, we assigned equivalently structured locus tags (with an “a,” “b,” “c,” etc., extension), the serial number being taken from the preceding ORF. Upon script-based checking for strict application of this rule, we detected four cases from NC_002607 in which the serial numbers of the “a”-extended codes were not taken from the preceding CDS. We decided to replace the serial number for these ORFs (see Table A4 in Appendix 1), reasoning that conflicts are excluded because these codes are no longer retrievable via RefSeq.
Correlation of protein-coding genes and spurious ORFs between strains R1 and NRC-1
The DNA sequence of the strain R1 and NRC-1 chromosomes is virtually identical (except for strain-specific transposon targeting) (Pfeiffer, Schuster, et al., 2008). The plasmids also show only few sequence differences in 350 kb of unique shared sequence (see Appendix 3) (Pfeiffer, Schuster, et al., 2008). However, nearly 25% of all the annotated ORFs which could be correlated showed start codon assignment discrepancies. All protein-coding genes with start codon assignment discrepancies had been subjected to extensive curation (Pfeiffer, Schuster, et al., 2008), taking into account extensive effort to validate the start codon assignments in strain R1 (Aivaliotis et al., 2007; Falb et al., 2006; Pfeiffer, Broicher, et al., 2008). We adjusted the NRC-1 genome annotation to that of the extensively curated strain R1. This included an effort for a totally consistent annotation of the duplicated plasmid regions in NRC-1. After this, we made sure that all closely related copies of a transposon have their transposase consistently annotated.
Whenever applicable, correlated protein-coding genes from strains R1 and NRC-1 are listed together (Tables S1–S6 (via Zenodo)). We also list strain-specific protein-coding genes from strain NRC-1 (Table S7 (via Zenodo)) and ORFs in GenBank (AE004437, AE004438, AF016485) which we consider to be spurious (Table S8 (via Zenodo)).
APPENDIX 6
PROTEIN FUNCTION ANNOTATION OF THE THREE ANALYZED HALOBACTERIUM STRAINS (91-R6, R1, AND NRC-1)
Gene and ORF annotation for strain NRC-1
We define the annotation of the strain R1 genome as reference annotation due to the significant efforts taken to ensure its reliability (Pfeiffer, Broicher, et al., 2008; Pfeiffer & Oesterhelt, 2015; Pfeiffer, Schuster, et al., 2008). The NRC-1 annotation was replaced by the annotation from strain R1 (protein name, gene, and EC number). Adequate handling was ensured for genes which are disrupted in only one of the strains. While spurious ORFs are not reported for strain R1 in GenBank, we retained spurious ORF annotations if the corresponding ORF is called in the current annotation of NRC-1 (AE004437, AE004438, AF016485) (Ng et al., 1998, 2000). For NRC-1 specific genes, we applied a simplified version of our reported annotation strategy (Pfeiffer & Oesterhelt, 2015; Pfeiffer et al., 2018).
The corrected annotation of the NRC-1 genome was submitted to NCBI as third party annotation (accessions: BK010829, chromosome; BK010830, plasmid pNRC100; BK010831, plasmid pNRC200).
Gene annotation for strain 91-R6
We define the annotation of the strain R1 genome as reference annotation (see above, annotation for strain NRC-1). All protein-coding genes from strains 91-R6 and R1 which occurred in matchSEGs had been correlated. In case of start codon assignment discrepancies, we rechecked the start codon assignment (because gene prediction by GenMarkS-2 is of high reliability and thus may uncover start codon assignment errors in R1). The annotation from the gene in strain R1 (protein name, gene, and EC number) was transferred to the gene from strain 91-R6. For 91-R6 specific genes, we applied a simplified version of our reported annotation strategy (Pfeiffer & Oesterhelt, 2015; Pfeiffer et al., 2018).
APPENDIX 7
THE DIVSEGS FROM STRAINS 91-R6 AND R1
For the three very long divSEGs (divSEG04, divSEG12, and divSEG18) see the main text. DivSEGs 03, 07, 09, 10, 13, 19, 25, 26, 27, 28, and 29 represent MGE insertions.
DivSEG02 is a 133 bp deletion in strain R1 in the rRNA operon promoter region (see also Appendix 9).
DivSEG05 corresponds to a replacement where the 91-R6 sequence is 1,537 bp and codes for an uncharacterized protein, carrying also a MGE remnant. Strain R1 has a 9,180 bp region with less than 60% GC which codes for a type I restriction enzyme (RmeMSR). The methyltransferase subunit RmeM has been targeted by a transposon. This is one of the transposons which occurs only in strain R1 but not in strain NRC-1, the latter strain thus coding for a functional restriction enzyme.
DivSEG22 and 23 are a 759 bp and a 246 bp deletion, respectively, in strain R1 and together cause disruption of the inosine-5′-monophosphate dehydrogenase paralog guaB2. The 3′ end of matchSEG21, the complete 57 bp matchSEG22, and the 5′ end of matchSEG23 code for this pseudogene remnant.
DivSEG27 is a 411 bp insert in strain 91-R6. It represents the integration of a MITE (MITEHsal2) into the N-terminal region of the bacteriorhodopsin (bop) gene. The protein sequences are identical for 259 residues but the N-terminal tripeptide Met-Leu-Glu of strain R1 is replaced by the tetrapeptide Met-Thr-Pro-Ser. The MITE insertion not only alters the signal sequence of the precursor protein but also the predicted stem-loop near the 5′ RNA (Srinivasan, Krebs, & Rajbhandary, 2006) and disconnects the CDS from the natural bop promoter. We are not aware of any studies showing that strain 91-R6 can produce a functional Bop or purple membrane.
DivSEG32 is a 1,475 bp indel which codes for a 2nd proline–tRNA ligase (proS2; HBSAL_10735) in strain 91-R6. This isoform shows only 23% protein sequence identity to proS1 (HBSAL_02515, corresponding to OE_1595F) and close homologs are found in few haloarchaeal genomes. A distinction between an insertion in strain 91-R6 or deletion in strain R1 is not possible, even though the C-terminal heptapeptide is encoded on matchSEG32, because this C-terminal region shows little conservation in the closest homologs.
DivSEG37 is a 891 bp deletion in strain R1 which removes an internal segment from a solo substrate-binding protein of an ABC transporter (OE_4225F).
DivSEG39 is a 5,311 bp deletion in strain 91-R6 which removes three poorly characterized genes, and most of the two subunits of a heterodimeric ribonucleoside-diphosphate reductase (nrdAB; OE_4346R + OE_4345R). Only a short C-terminal remnant of the beta subunit is retained (HBSAL_11665). This gene pair is not essential as both strains also code for a distantly related (25% protein sequence identity) monomeric enzyme (nrdJ; subunits fused; OE_3328R; HBSAL_08550).
Several divSEGs code for integrases or integrase domain proteins. In two cases, there are tRNA genes at or close to the integration point (divSEG15, divSEG30). In two cases, the divSEG has targeted a protein-coding gene and is bounded by direct repeats (divSEG14, divSEG31).
DivSEG14 is a 3,244 bp insert in strain 91-R6 which has targeted a NamA family protein (OE_2360R, HBSAL_05570 + HBSAL_05545). It codes for an integrase family protein and three uncharacterized proteins.
DivSEGs 15, 16, and 17 are all below 60% GC, and are separated by very short matchSEGs (634 bp, 901 bp). In R1, DivSEG15 is flanked by a tRNA-Lys gene at its left end and by a 27 bp direct repeat of the 3′ end of the tRNA-Lys at its right end. It has a 9,530 bp region (only in R1), and codes for two MGEs, six genes without well-defined function and an integrase located at the extreme right end. The overall arrangement is typical of integrative elements, such as a provirus, that has targeted a chromosomal tRNA gene. DivSEG16 (1,197 bp only in R1) codes for an ORFan. In divSEG17, a 1,086 bp region of R1, coding also for an ORFan, is replaced by a 3,215 bp region of strain 91-R6 which carries a MGE remnant and a gene without well-defined function. At the junction to the subsequent matchSEG, a homolog to a phiH1-like repressor protein is encoded. Altogether, this gives the impression of a provirus remnant.
DivSEG30 is a 7,561 bp insert in strain 91-R6 which targets and thus duplicates a tRNA-Gly. It codes for an integrase family protein and 10 uncharacterized or only generally characterized proteins.
DivSEG31 is a 4,839 bp insert in strain 91-R6. This has targeted a GNAT acetyltransferase domain protein (R1: OE_3592F; 91-R6: N-terminal part HBSAL_09405, C-terminal part HBSAL_09440). The insert codes for an integrase family protein and five genes without well-defined function.
APPENDIX 8
JUNCTION ANALYSIS AT THE TERMINI OF CORRESPONDING REGIONS ON THE PLASMIDS FROM STRAINS 91-R6 AND R1
The termini of high similarity regions represent strain-specific junctions which may allow the parental sequence to be delineated. Junction analysis thus may reveal the evolutionary history and processes.
Junction JA1
We assigned junction JA1 to the 5′ end of the 39.2 kb duplication between pHSAL1 and pHSAL2 (Figure A2 in Appendix 2). At this end of the duplication is an ISHsal4 transposon which has been targeted by transposon ISH5. At this junction, the parental sequence cannot be delineated as a TSD is lacking in both plasmids and there are no targeted genes.
Junction JA2
We assigned junction JA2 to the 3′ end of the 39.2 kb duplication between pHSAL1 and pHSAL2 (Figure A2 in Appendix 2). At this end of the duplication is an ISH1 transposon. The junction traverses the point of ring opening of both plasmids. At this junction, pHSAL2 can be unambiguously discerned as the parent. The sequence upstream of ISH1 is repeated as a TSD in pHSAL2 (AGCCGCCA). Additionally, a pseudogene upstream of the transposon (HBSAL_13535; HBSAL_12845) is homologous to the N-terminal region (ca amino acids 1–150) of C481_14553. A homolog to the C-terminal region (ca amino acids 150–620) is encoded only on pHSAL2 (HBSAL_13005).
Junction JB1
We assigned junction JB1 to a contiguous sequence in R1 plasmid pHS3 which matches to disconnected and oppositely oriented regions on the chromosomal strain-specific divSEG12 of strain 91-R6 (p3I = c16; p3J = c10; Figure 2). At both involved junctions in strain 91-R6 is a MGE of subtype ISH3C. The match overlaps by 5 bp in pHS3 (ATGAT), which is indicative of a 5 bp TSD, typical for ISH3-type transposons. This is best explained by pHS3 representing the parental sequence, and the sequence in divSEG12 having become rearranged. Population heterogeneity involving this pair of ISH3C elements was encountered (see Figure 5 and Appendix 10). It should be noted that the transposon of subtype ISH3B, which is located upstream of region c10, participates in junction JB2 on its other side.
Junction JB2
We assigned junction JB2 to a contiguous sequence on divSEG12 (c10/c11; Figure 2). In R1 plasmid pHS3 are multiple transposons and one strain-specific sequence. The two transposons of subtype ISH8B lack a TSD in the current configuration, but in combination there are “hybrid TSDs” on both sides (AGTCGTATCC and CTTCGAGGCGG) (Figure 6). This supports the assignment of plasmid pHS3 as being inverted at this junction. Support comes from a split pseudogene adjacent to this ISH8B element pair. Combined, OE_5405F and OE_5013R correspond to HBSAL_04690 and are a close full-length homolog of Halxa_0005. Further support comes from a split transposon (ISH32), the fragments of which occur on the other side of the ISH8B element pair and, combined, correspond to a complete MGE, including a hybrid TSD (GGAGGGCGGG) (Figure 6).
Junction JB3
We assigned junction JB3 to the boundaries of a strain-specific 8 kb sequence in divSEG12 from strain 91-R6. The fact that the sequence in divSEG12 is consecutive with the adjacent sequences without intervening MGEs supports the view that this is the parental configuration. This is supported by the lack of a TSD at the equivalently positioned ISH2 element in R1 plasmid pHS3. Further support comes from the pseudogene OE_5019R which is truncated at the ISH2 element and corresponds to the N-terminal region of the regular protein-coding gene HBSAL_04810. This is best explained by pHS3 having been targeted independently by two copies of ISH2 with subsequent recombinations, so that one copy and the intervening sequence have been lost.
Junction JC1
We assigned junction JC1 (Figure 3) to correlated but independently disrupted homologs of ACP99_RS08965 (WP_049986279.1). The R1 homolog OE_5394R is encoded on pHS3 and is disrupted by an ISH2 element which is bounded by an extremely long TSD (55 bp). This causes duplication of 18 codons. In strain 91-R6 the multiply-disrupted gene is targeted by a transposon of subtype ISH3B. The N-terminal region (HBSAL_05030) is encoded in the strain-specific region divSEG12 (region c16; Table 6) of the chromosome, which terminates with an ISH3B transposon. The central and C-terminal parts, additionally targeted by another MGE (ISHsal2), are encoded in the duplicated part of plasmids pHSAL1/pHSAL2 (HBSAL_12805 + HBSAL_12815; HBSAL_13495 + HBSAL_13505). They are encoded downstream of a transposon of subtype ISH3B (which in turn has been targeted by transposon ISH5). The chromosomal and plasmid copies of ISH3B exemplify a hybrid TSD (AAATT), indicative of an MGE-triggered genome rearrangement.
Junction JC2
We assigned junction JC2 (Figure 4) to homologs of rrnAC2017 from Har. marismortui, a protein which has multiple full-length homologs in other haloarchaeal genomes. Homologous to the N-terminal region (ca amino acids 1–50) is a pseudogene in the strain-specific segment divSEG12 on the chromosome (HBSAL_04640; encoded on c09; Table 6). Homologous to the C-terminal region (ca amino acids 51–249) is a pseudogene pair which is encoded on the 39.2 kb duplication between pHSAL1 (HBSAL_12720) and pHSAL2 (HBSAL_13410). These junctions are not immediately adjacent to MGEs.
APPENDIX 9
USING THE STRAIN 91-R6 CHROMOSOME SEQUENCE AND DATA FROM A NRC-1 RESEQUENCING PROJECT TO INTERROGATE THE CHROMOSOMAL DIFFERENCES BETWEEN STRAINS R1 AND NRC-1
- Four individual sequence corrections between nt 30,407 and 30,520 of the chromosome, which are within transposon ISH1, make the NRC-1 sequence identical to the R1 sequence.
- At three positions of the chromosome, polynucleotide runs were shortened (nt 425,429, nt 460,883, nt 586,819). In all three cases, this makes the NRC-1 sequence identical to the R1 sequence.
- A point mutation at nt 1,023,692 makes the NRC-1 sequence identical to the R1 sequence.
- Insertion of a C at nt 1,230,902 makes the NRC-1 sequence identical to the R1 sequence.
- A point mutation at nt 271,110 of pNRC200, which is within transposon ISH6, makes the NRC-1 sequence identical to the R1 sequence.
- At position 3,393 of pNRC100 and pNRC200, a point mutation was found. Here, the original sequence of NRC-1 corresponds to that of strain R1.
- Three novel target sequence duplications were detected in the chromosome, implying integration of a further transposon copy. In all three cases, the original NRC-1 sequence corresponds to that of strain R1.
- A 24.2 kb deletion was detected in pNRC200. This region is present in R1 plasmid pHS3 (pos 9,767–21,755, which is part of region p3A, see Table 5).
Aside from differences related to ISH elements, the chromosomes of strains R1 and NRC-1 (as originally published) show only 12 differences: four point mutations, five single-base frameshifts, and three indels (Pfeiffer, Schuster, et al., 2008).
Indel differences
(a) Strain R1 has a 133 bp deletion in the rRNA promoter region (divSEG02) (Pfeiffer, Schuster, et al., 2008). The strain 91-R6 sequence corresponds to that of strain NRC-1. (b) Strain NRC-1 has a 423 bp deletion in hcyB (halocyanin), which removes one of two copper-binding domains (Pfeiffer, Schuster, et al., 2008). The type strain sequence corresponds to that of strain R1. (c) There is a 10,007 bp extra sequence with an 8 bp terminal duplication in strain NRC-1 compared to R1 (Pfeiffer, Schuster, et al., 2008). The strain 91-R6 genome matches that of strain R1. The insertion occurs in the center of the pilB2 gene, which is thus disrupted in strain NRC-1. The adjacent pilC2 gene is disrupted by an in-frame stop codon in all three strains, consistent with previous observations (Losensky et al., 2015; Pfeiffer, Schuster, et al., 2008). With its partner gene defective, the pilB2 gene of R1 is probably without function even though not being disrupted itself. The 10,007 bp region from NRC-1 has proviral characteristics (having integrase and phage primase related genes). It has been discussed that this could be an insertion in NRC-1 which occurred after the branching of R1 and NRC-1. Alternatively, the insertion occurred in the ancestor of R1 and NRC-1 but with a subsequent repeat-mediated deletion in R1 (Dyall-Smith et al., 2011). The strain 91-R6 sequence corresponds to that of strain R1, thus making it more likely that the 10,007 bp sequence is an insertion in strain NRC-1.
Frameshift differences
Of the five frameshift differences between strains R1 and NRC-1, four are within coding regions. (a) OE_1823F is identical to HBSAL_03125 at the DNA sequence level. The frameshifted NRC-1 protein VNG_0553C is annotated as a regular protein but has a long C-terminal region which overlaps with the coding region of VNG_0553a/OE_1827F/HBSAL_03130. The resequenced NRC-1 genome does not have a frameshift and corresponds to the R1 sequence. (b) OE_1916F is identical to HBSAL_03355 at the DNA sequence level. The frameshifted NRC-1 protein VNG_0606G is disrupted. The resequenced NRC-1 genome does not have a frameshift and corresponds to the R1 sequence. (c) OE_2141F is identical to HBSAL_04035 at the protein sequence level. The frameshifted NRC-1 protein VNG_0779C is disrupted. The C-terminal part was initially annotated as VNG_0780H. The resequenced NRC-1 genome does not have a frameshift and corresponds to the R1 sequence. (d) OE_3338R is identical to HBSAL_08590 at the DNA sequence level. In NRC-1, the gene is affected by a frameshift, which does not occur in the resequenced NRC-1 genome. In addition to that initial frameshift difference, the gene also differs by having been targeted by two transposons. The C-terminal part is annotated as regular protein VNG_1650H, with a hybrid start codon, the first two bases of which are part of a targeting transposon.
APPENDIX 10
POPULATION HETEROGENEITY IN STRAIN 91-R6
The genomic heterogeneity of Hbt. salinarum strain 91-R6 could be analyzed in detail using PacBio long sequencing reads. All heterogeneities were found to be associated with mobile genetic elements (MGEs). PacBio reads were assigned as representing distinct isoforms by BLASTn analysis. We selected unique regions (typically 150 bp) adjacent to such MGEs, joined them into one contiguous sequence, and used the BLASTn results to determine the connectivity of the individual PacBio reads. In the case of an optional MGE (i.e. one that is present in only part of the population), BLAST hits may either be contiguous (if the MGE is lacking) or may be noncontiguous, with a gap reflecting the length of the MGE. Optional MGEs were encountered for transposons (ISHsal1 and ISHsal15), as well as for a MITE (MITEHsal2). In the case of a genome rearrangement, a PacBio read would show BLASTn hits to unique regions adjacent to distinct copies of the MGE. If the BLASTn hit pattern did not allow classification, we extended the query to include the complete PacBio read and used it to search (BLASTn) against the assembled genome for a more detailed analysis.
The most prominent heterogeneities were identified in four regions. Genome rearrangements were encountered only in two regions, both of which are located in divSEG12, which is the 164 kb strain-specific, plasmid-like sequence in strain 91-R6 which replaces a 2,306 bp region from strain R1. In the following, heterogeneities are described in order of increasing complexity.
Case A
Optional copies of the transposon ISHsal1 and MITE, MITEHsal2, were identified. These were separated by 14.6 kb (MITEHsal2 integrated at nt 935,890–935,896 in reverse orientation with 7 bp TSD TAAGCCA; ISHsal1 integrated at nt 950,574–950,578 with 5 bp TSD, AGTAT). In both cases, we selected the version lacking the transposon for the representative genome. With respect to MITEHsal2, 405 PacBio reads confirmed the assembly, with slightly more reads being contiguous instead of having the inserted MITE (Figure 7). There were 10 PacBio reads which indicated MITEHsal2 triggered genome rearrangements, with six distinct connections. With respect to ISHsal1, 415 PacBio reads confirmed the assembly, with more reads being contiguous instead of having an inserted transposase (Figure 8). In addition, we encountered 58 PacBio reads representing ISHsal1 triggered genome rearrangements, with five different connections. About half of these indicated the integration of plasmid PHSAL2 into the chromosome. There were 17 PacBio reads which covered both the MITEHsal2 and the ISHsal1 heterogeneity (Figure 9). Among these, eight reads lacked both transposons, five contained both, and two contained only one of the MGEs (MITEHsal2). The remaining two showed genomic rearrangements over one of the MGEs. The reads having only a single MGE indicate that integration of MITEHsal2 preceded integration of ISHsal1 (Figure 9). It should be noted that an independent optional copy of MITEHsal2 was encountered elsewhere in the genome (see below, case D).
Case B
In the chromosome, we found a 23.8 kb inversion which is bounded by oppositely oriented copies of transposon ISHsal1 (Figure 8). The orientation which we selected for the representative genome is supported by a targeted pseudogene with traverses one of the elements (HBSAL_04465 and HBSAL_04475) and contains a target site duplication (AGTTT) for one of the copies. This version also has a slightly higher coverage by PacBio reads. In addition to the 577 reads which confirmed the assembly over one or the other of the two junctions, we encountered 133 PacBio reads which represented additional genome rearrangements. Such rearrangements were detected for all other copies of transposon ISHsal1, including the copy on plasmid PHSAL2, which thus means that the plasmid has been integrated into the chromosome in these cases (see also case A for an equivalent observation).
Case C
The representative genome contains a single, complete copy of ISHsal15 near position 851 kb, within the 164 kb plasmid-like sequence (divSEG012) that is specific for strain 91-R6. A second, optional copy was encountered at 1,054 kb, which is 202.6 kb away from the first copy, and is found within matchSEG14 (Figure 10). There were 259 PacBio reads that lacked the optional copy of ISHsal15, and 19 reads that contained it. Curiously, PacBio reads supporting a ISHsal15-triggered 202.6 kb genome inversion were much more frequent (45 and 53 reads traversing the two ends, respectively). The ISHsal15 copy at 851 kb has been partially deleted together with an adjacent 16 kb region (see case D).
Case D
Several rearrangements were identified that were associated with copy 2 of transposon ISH3C (nt 868,513–869,901, forward orientation). The genome contains four copies of ISH3C, all of them within the 164 kb plasmid-like sequence specific for strain 91-R6 (divSEG12). In the representative genome, copies 2 and 4 (nt 925,785–927,173, reverse orientation) are identical in sequence, oppositely oriented, and 55.8 kb apart. A genome inversion triggered by these copies of ISH3C was encountered. The version assumed to be parental is supported by 161 PacBio reads traversing copy 4 while the inverted version, which was selected for the representative genome, is traversed by 74 reads. On the other side of ISH3C copy 2 is a 16 kb sequence, terminating with ISHsal15 (see above, case C). A deletion covers this 16 kb sequence, including a short region from ISHsal15. While 91 reads traverse ISH3C copy 2 in the version selected as representative, only 12 cover it in the inverted version, assumed to be parental and still containing the 16 kb sequence. The version lacking the 16 kb sequence is supported by 144 PacBio reads, which indicates a strong drift toward removal of that sequence. Deletion of the 16 kb sequence also eliminated the only copy of ISHsal16 from the genome. In addition, that sequence contained a nonoptional as well as an optional copy of MITEHsal2 (absent on 142 reads, present on 68 reads). There are six PacBio reads which contain the 51 kb region between copies 2 and 4 of ISH3C in inverted orientation but lack the 16 kb sequence. This is attributed to an independent inversion of the genome region subsequent to deletion of the 16 kb sequence. Copies 1 (nt 811,634–813,022, forward orientation) and 3 (nt 901,476–902,864, reverse orientation) of ISH3C are identical to each other and show 96% DNA sequence identity to copies 2/4. Their relative orientation depends on the orientation of the invertible 55.8 kb sequence. A few additional genome rearrangements were encountered in a low number of reads, including deletions due to rearrangements between identically oriented copies of ISH3C, likely to reflect the parental sequence.
APPENDIX 11
MGE ANALYSIS
This text provides additional details of MGE analysis, including definitions, nomenclature issues, and special cases. We adopted the standards defined by ISFinder (Siguier et al., 2012).
MGEs of type transposon
Transposons are mobile elements which encode their own transposase for mobilization. Commonly, transposons contain an inverted terminal repeat. We refer to transposons with that characteristic as “canonical transposon.” Typically, multiple copies of the same transposon in a genome are extremely similar to each other, if not identical. We refer to the integration of a strain-specific copy of a transposon as “transposon targeting.” Such events are detected as an indel upon genome alignment. Many divSEGs detected upon chromosome comparison reflect transposon targeting (see Appendix 7). In plasmid comparisons, correlated sequences may terminate at a transposon targeting site.
Transposase sequences are much better conserved on the protein level than the transposon DNA sequences. Based on transposase homologies, transposons can be grouped at higher levels. We have assigned transposons to classes on an ad-hoc basis and have grouped our results according to transposon class.
Halobacterium also contains several “noncanonical” transposons (ISH7, IS605-type, the latter being the combination of IS200-type and IS1341-type). For these transposons, attempts to pinpoint the termini may fail, which complicates analyses. For reasons of simplicity we skipped noncanonical transposons in our analyses, since none of the strain differences were related to such MGEs. However, noncanonical transposons are fully covered in the annotation of the genome.
MGEs of type MITE
MITE stands for “Miniature Inverted-Terminal-repeat Element.” MITEs are mobilized in trans by transposases encoded on transposons. For this to be possible, the inverted terminal repeats of the MITE and the associated transposon have to be homologous to each other. This is also the basis for assignment of a MITE class. The collection of MITEs in ISFinder has started only recently.
In Halobacterium, one MITE is known since long (ISH2) but is typically referred to as transposon even though it does not code for a transposase. ISH2 codes for a short protein which has been identified by proteomics (Klein et al., 2007). In ISFinder, ISH2 is integrated into the transposon section and not into the MITE section.
Transposon assignment and naming
According to ISFinder, MGEs which show 95% DNA sequence identity are considered the same transposon, even if they occur in distinct organisms. We have adopted this principle. Historically, a transposon name for Halobacterium consists of the term ISH, followed by a serial number (e.g. ISH1, ISH4, ISH6). The elements which were historically described as ISH3 and ISH8 are diverse and would now be considered distinct transposons according to current ISFinder principles. We resolve this by addition of a letter (ISH3B, ISH3C, etc.; ISH8A, ISH8B, etc.).
One transposon (ISNpe8) is closely related to ISH10 and thus was initially not considered a distinct MGE in the laboratory strains of Halobacterium. This variant was detected in Natrinema pellirubrum and was submitted to ISFinder under a name based on that species. Because the element in Halobacterium is near-identical, it has to be listed under that “foreign” name. Additionally, we detected a copy of ISNpe16 in strain 91-R6 which is in ISFinder under that name.
When a considerable number of novel transposons were detected in strain 91-R6, it had to be decided if the historical naming convention for Halobacterium should be continued (which would have resulted in large serial numbers) or if the novel elements should follow current naming conventions. Together with ISFinder, it was decided to adopt current naming conventions and to name novel transposons from strain 91-R6 with prefix ISHsal, followed by a serial number. All novel transposons from strain 91-R6 were integrated into ISFinder with names based on this principle.
Finally, there are transposons which are complete by our definition (both termini are intact without long internal deletions). However, the transposase gene of these MGEs is disrupted which makes them unsuitable for ISFinder. Typically, we attempted to identify a homologous element in another genome which is complete and carries a nondisrupted transposase gene, and to submit that to ISFinder so that a regular name is assigned. If an ISFinder-compatible element cannot be identified, we process these elements as “HsIRS” (Halobacterium salinarum ISH-Related Sequence). Only a few of the annotated HsIRS are canonical and complete, and these are included in our analyses.
MITE assignment and naming
As MITEs became more closely studied well after transposons, even Halobacterium MITEs had not been examined in detail. The exception was ISH2, which has been processed as an atypical type of transposon and thus has been given its historic name. All other MITEs were only recently annotated by us and have received a name which is consistent with current ISFinder rules (prefix MITEHsal followed by a serial number).
Attempts to identify the potential source of a MGE
Attempts to identify a potential source were made only for MGEs which are specific for the type strain (91-R6) or for the laboratory strains (R1 and NRC-1). Plasmids and plasmid-like sequences typically carry many MGEs. If a plasmid-like sequence is taken up by a cell and the plasmid either manages to multiply as an episome or to integrate into the chromosome of its novel host, this may lead to “infection” with the set of MGEs which are carried along. In the most simple scenario, the MGE is retained in the genome within the context of is “original source.” For an MGEs with a single copy, the currently occupied genome region is assigned as its potential source. By this scheme, many MGEs are assigned to the long strain-specific sequences (divSEG04, divSEG12, divSEG18) and those plasmid regions which are not shared between type and laboratory strains.
For MGEs with multiple copies, a more elaborate analysis is required. Events of transposon targeting are considered to represent mobilization events, excluding them to be classified as the original source. For R1, transposon targeting can be detected not only by comparison to the type strain, but also by comparison to NRC-1. In several cases, all but one copy showed a signature of transposon targeting and thus that copy was assigned as a potential source. If more than one copy was not involved in targeting, we list more than one potential source.
For one case each in strain 91-R6 and R1, all copies showed a signature of transposon targeting. In this case, we classified the potential source as “unknown.”
Open Research
DATA AVAILABILITY STATEMENT
The nucleotide sequence accession numbers for Hbt. salinarum 91-R6 (DSM 3754T) in GenBank are CP038631 (chromosome), CP038632 (plasmid pHSAL1), and CP038633 (plasmid pHSAL2). The Third Party Annotation accession numbers for Hbt. salinarum NRC-1 are BK010829 (chromosome), (BK010830) (plasmid pNRC100), and BK010831 (plasmid pNRC200). Supplementary Tables S1–S6 have been uploaded into a single PDF file at Zenodo (https://doi.org/10.5281/zenodo.3528126).