High-quality genome of a novel Thermosynechococcaceae species from Namibia and characterization of its protein expression patterns at elevated temperatures
Nathanael D. Arnold and Michael Paper contributed equally to this work.
Graphical Abstract
In this study, the genome of a new thermophilic cyanobacterium, Thermosynechococcaceae cyanobacterium sp. Okahandja, isolated from a hot spring near Okahandja in Namibia, was sequenced. Additionally, cultivations were conducted at elevated temperatures of 40, 50, and 55°C, followed by analyses of the respective adapted proteomes based on the annotated genome.
Abstract
Thermophilic cyanobacteria thrive in extreme environments, making their thermoresistant enzymes valuable for industrial applications. Common habitats include hot springs, which act as evolutionary accelerators for speciation due to geographical isolation. The family Thermosynechococcaceae comprises thermophilic cyanobacteria known for their ability to thrive in high-temperature environments. These bacteria are notable for their photosynthetic capabilities, significantly contributing to primary production in extreme habitats. Members of Thermosynechococcaceae exhibit unique adaptations that allow them to perform photosynthesis efficiently at elevated temperatures, making them subjects of interest for studies on microbial ecology, evolution, and potential biotechnological applications. In this study, the genome of a thermophilic cyanobacterium, isolated from a hot spring near Okahandja in Namibia, was sequenced using a PacBio Sequel IIe long-read platform. Cultivations were performed at elevated temperatures of 40, 50, and 55°C, followed by proteome analyses based on the annotated genome. Phylogenetic investigations, informed by the 16S rRNA gene and aligned nucleotide identity (ANI), suggest that the novel cyanobacterium is a member of the family Thermosynechococcaceae. Furthermore, the new species was assigned to a separate branch, potentially representing a novel genus. Whole-genome alignments supported this finding, revealing few conserved regions and multiple genetic rearrangement events. Additionally, 129 proteins were identified as differentially expressed in a temperature-dependent manner. The results of this study broaden our understanding of cyanobacterial adaptation to extreme environments, providing a novel high-quality genome of Thermosynechococcaceae cyanobacterium sp. Okahandja and several promising candidate proteins for expression and characterization studies.
1 INTRODUCTION
Cyanobacteria are a diverse group of photosynthetic microorganisms that have significantly influenced the evolution of our planet through their impact on global carbon and nitrogen cycling, enabling life for oxygen-dependent species (Knoll, 2008). Among them, thermophilic cyanobacteria have emerged as fascinating model organisms that thrive in extreme habitats characterized by high temperatures (Ward et al., 2012). They possess unique molecular and physiological adaptations, allowing them to adapt to environments where most other organisms cannot survive. Known mechanisms that confer thermophile cyanobacteria their ability to thrive in these adverse environments include cell membrane lipid composition alterations (homoviscous and homeophasic adaptation), higher GC content in rRNAs and tRNAs compared to mesophilic bacteria (Galtier & Lobry, 1997), changes in protein structure with more intramolecular salt bridges and fewer polar lipids per surface area (Thompson & Eisenberg, 1999), and potential production of more naturally heat-resistant macromolecules as opposed to low MW molecules. Additionally, smaller genome sizes are a ubiquitous phenomenon for thermophilic organisms (Sabath et al., 2013; Van Noort et al., 2013), while functional simplification appears to be a common genomic adaptation mechanism in thermophilic prokaryotes (Burra et al., 2010).
Thermophilic microorganisms and their inherently thermostable enzymes are highly relevant for biotechnological applications since they allow for unsterile culture conditions without the threat of microbial contaminations, additionally requiring less energy-intense cooling (Patel et al., 2019). In this context, the product spectrum of (thermophilic) cyanobacteria includes pigments (Graverholt & Eriksen, 2007), vitamins (Abed et al., 2009), poly-β-hydroxybutyrate (PHB) (Nishioka et al., 2001), bioactive compounds (Fish & Codd, 1994; Kumar et al., 2019; Singh et al., 2011), or lipids (Bywaters & Fritsen, 2015; Maslova et al., 2004). Further promising application fields comprise nutritional supplements (Watanabe et al., 1999), biofertilizers (Kumar et al., 2019), wastewater clearing (Aksu et al., 2009; Ertuǧrul et al., 2008; Sawayama et al., 1998), CO2 fixation (Hsueh et al., 2007; Leu et al., 2013), biofuel production (Karatay & Dönmez, 2011), or rare earth recycling (Paper et al., 2023). Despite their ubiquity and promising biotechnological applications, relatively little is known about the differential protein expression of cyanobacteria in response to elevated temperatures. While several studies and reviews (Tang et al., 2022; Wang et al., 2015) engaged with this scientific problem with the example of thermophilic bacteria (Chen et al., 2013; Gao et al., 2004; Pysz et al., 2004; Shih & Pan, 2011; Trauger et al., 2008; Wang et al., 2012), more research specifically aimed at cyanobacteria is required.
Cyanobacteria can be found in habitats with a wide range of environmental conditions (Callieri et al., 2019; Oren, 2015). Due to their geologically separated nature, hot springs are relevant island-like ecosystems for speciation events, driven by local community dynamics (Ionescu et al., 2010). Members of the genus Thermosynechcococcus have been documented in hot springs with temperatures ranging from 44°C to 98°C (Saxena et al., 2017; Stolyar et al., 2014; Tang et al., 2018; Ward et al., 2017). Given their limited evolutionary distance, they give insights into how rapid environmental adaption can be achieved.
The investigated cyanobacterial strain of this study was isolated from a hot spring near the city of Okahandja in Namibia. We aimed to gain a comprehensive system biology-driven understanding of the temperature adaptation of this newly isolated strain. Therefore, a multidisciplinary approach was deployed, combining PacBio-guided long-read genome sequencing and high-resolution nLC-Q-TOF mass spectroscopy-driven proteome analyses to unravel the distinct genetic features and metabolic adaptations that allow this thermophilic cyanobacterium to thrive in environments characterized by elevated temperature regimes. Through comparison of our high-quality genomic data and the intracellular proteomes derived from cultures grown at different temperature profiles, we identified key genes and metabolic pathways involved in the adaptation to extreme temperatures and discussed the results in the context of comparable studies. In addition, sequence alignment with the closely related strain Parathermosynechococcus lividus PCC 6715 revealed extensive gene flux and rearrangements.
Maximum likelihood analysis based on 16S rRNA gene sequences and the concatenated phylogenetic tree indicated allopatric speciation, which was reinforced by ortholog-based phylogenetics of Thermosynechococcaceae. Accordingly, the novel specimen was placed close to the branch of the recently defined genus of Parathermosynechococcus, which incorporates the Yellowstone National Park-originating P. lividus PCC 6715 (Tang et al., 2024). Based on these combined findings, we suggest the name Thermosynechococcaceae cyanobacterium sp. Okahandja for the novel thermophilic cyanobacterium presented in this study.
2 MATERIALS AND METHODS
All chemicals were retrieved from Sigma-Aldrich, while VWR supplied general consumables. All necessary buffers and enzymes for next-generation genome sequencing were shipped from Pacific Biosciences. High-molecular-weight DNA was extracted using the Quick-DNA™ HMW MagBead Kit from Zymo Research. HMW gDNA shearing was conducted using g-TUBEs (Covaris). Polymerase chain reactions were done using the DreamTaq Polymerase and corresponding buffer (Thermo Fisher Scientific). For agarose gel DNA extraction, the Monarch DNA Gel Extraction Kit (New England BioLabs GmbH) was utilized according to the manufacturer's protocol.
2.1 Cultivation
The cyanobacteria were isolated from a hot spring near Okahandja in Namibia (Callieri et al., 2019) and cultivated in an Infors Lab5 stirred photobioreactor system (Infors HT) in BG11 medium according to Stanier et al. (1971) at temperatures of 40°C, 50°C, and 55°C. Stirring speed was set to 150 rpm and the illumination intensity was 200 µmol m−2 s−1 PPFD. During cultivation, the culture was aerated with 1.016 Nl/min through a sparger. By adjusting the in-gas CO2 portion, the pH was maintained at 8.0. For proteome analysis, the biomass was harvested during the exponential growth phase after 7 days.
2.2 Microscopy
Microscopic images were taken using a confocal laser scanning microscope LSM 980 (Carl Zeiss) equipped with an Axiocam 508 color imaging system. Secondary electron (SE) and transmission electron images were taken using a JSM-7500F Field Emission Electron Microscope (Jeol Ltd.) equipped with a low-energy ionization detector (LEI) and a transmission electron detector in scanning mode (STEM). The SE images were taken with an accelerating voltage of 1 kV and the STEM images were taken with 30 kV. Before sample preparation, the culture was washed with demineralized water to remove salts.
2.3 Molecular biology analysis
2.3.1 Species identification
16S ribosomal RNA gene-inferred phylogenetic analysis
The full-length 16S ribosomal RNA gene sequence was extracted from the whole-genome nucleotide sequence using Barrnap (Seemann, 2013), as implemented on the public Galaxy.eu server of the Galaxy web platform (Afgan et al., 2016). Using the BLAST tool of the National Center for Biotechnology Information (NCBI) GenBank, the 16S rRNA gene sequence was compared to already submitted sequences of cyanobacterial strains from public culture collections in terms of authenticity. The assembled 16S rRNA gene sequence and related sequences of cyanobacterial strains cited from GenBank were used for phylogenetic analysis, including Gloeobacter spp. as an outgroup. For the 16S rRNA gene alignment, the Muscle algorithm in Mega X (Kumar et al., 2018) was applied. The phylogenetic tree was calculated in Mega X using the Maximum Likelihood method (ML) and the General Time Reversible model (Nei & Kumar, 2000) with Gamma-distributed rates and Invariant sites (GTR + G + I model) and a bootstrap value of 1000. For the Bayesian phylogenetic analyses, two runs of eight Markov chains were executed for one million generations with default parameters with Mr. Bayes 3.2.1 (Ronquist & Huelsenbeck, 2003) as described in Jung et al., 2024 (Jung et al., 2024).
Orthology-inferred phylogenetic analysis
Using the public Galaxy.eu server of the Galaxy web platform (Afgan et al., 2016), OrthoFinder 2.5.4 (Emms & Kelly, 2015) was utilized to identify orthologs among the genomes of Thermosynechococcaceae as conducted by Jahodářová et al. (2022). Gene sequence searching was carried out using DIAMOND 2.0.15 (Buchfink et al., 2014) to find orthogroups (groups of genes descended from a single gene in the last common ancestor of a group of species). Unrooted gene trees were then inferred using DendroBLAST (Kelly & Maini, 2013). OrthoFinder utilizes STAG (Emms & Kelly, 2018) on default to infer unrooted species trees from all genes from the set of unrooted gene trees. These unrooted species trees are then rooted utilizing STRIDE (Emms & Kelly, 2017), from which OrthoFinder infers orthologs in a final step. See Emms and Kelly (2015) for details on the orthogroup inference stage and Emms and Kelly, (2019) for details on the second stage from orthogroups to gene trees, the rooted species tree, orthologs, gene duplication events, and so forth. In total, OrthoFinder assigned 85,177 genes (95.9% of the total) to 7952 orthogroups. Fifty percent of all genes were in orthogroups with 22 or more genes (G50 of all genes was 22) and were contained in the largest 1508 orthogroups (O50 was 1508). There were 451 orthogroups present within all compared Thermosynechococcaceae species' genomes, and 271 of these consisted entirely of single-copy genes (exactly one from each compared species). These 271 single-copy orthologs were utilized to infer the outgroup-free, phylogenetic species tree depicted in Figure 4. iTol (Letunic & Bork, 2024) was deployed to visualize the Newick formatted phylogenetic species tree acquired from OrthoFinder.
2.3.2 Next-generation genome sequencing
HMW genomic DNA extraction and DNA library preparation
About 50 mL of cyanobacterial culture was collected during the exponential growth phase and centrifuged at 10.000 rcf for 5 min. The biomass was used for the extraction of high-molecular-weight genomic DNA with the Quick-DNA HMW MagBead Kit (Zymo Research) following the manufacturer's instructions. Light absorption ratios at 260/280 nm wavelength were measured using a photometer (Nano Photometer NP80; IMPLEN) and a Qubit 4 fluorometer with the Qubit 1X dsDNA HS Assay-Kit (Thermo Fisher Scientific) to evaluate the purity and concentration of the obtained genomic DNA. Fragment sizes of the gDNA were analyzed using a FemtoPulse capillary electrophoresis instrument (Agilent Technologies). Samples that passed the quality control were then sheared using g-TUBEs (Covaris) with 1700 g in a tabletop centrifuge, resulting in DNA fragments of approximately 12 kb in size, as verified by FemtoPulse again. Subsequently, a HiFi library was then generated by following the instructions in the manual of the SMRTbell prep kit 3.0 (Pacific Biosciences), which involves attaching barcoded adapters to the gDNA fragments. The libraries were stored at −20°C until the day of sequencing, when the Sequel II Binding Kit 3.2 (Pacific Biosciences) was used to bind primers and polymerase to the samples, following the manufacturer's recommendations.
Sequencing
A single SMRT cell (lot number 418096) was used to perform whole-genome sequencing on a Sequel IIe platform (Pacific Biosciences) utilizing the following parameters: 2 h of pre-extension, 2 h of adaptive loading (with a target of p1 + p2 = 0.95) to obtain a final on-plate concentration of 85 pM, and a movie window of 30 h for signal detection (Saxena et al., 2017).
Assembly and annotation
Obtained FASTQ raw read files were assembled utilizing the Canu assembler 2.0 (Tang et al., 2018) with a user-provided estimated genome size of 2.7 Mbp (genomeSize = 2.7 mb) and the -pacbio parameter. The corresponding assembly report file can be found in the supplementary data at https://zenodo.org/doi/10.5281/zenodo.10007199. Gene prediction and annotation were performed using NCBI's Prokaryotic Genome Annotation Pipeline (PGAP) (Seemann, 2013; Stanier et al., 1971; Stolyar et al., 2014; Tang et al., 2024), comprising GeneMarkS-2+ for gene prediction (Afgan et al., 2016) and TIGRFAMs for functional identification of proteins (Jung et al., 2024; Kumar et al., 2018; Nei & Kumar, 2000; Ronquist & Huelsenbeck, 2003). Secondary annotation was performed using the browser-based tool RAST (Rapid Annotation using Subsystem Technology) (Buchfink et al., 2014; Emms & Kelly, 2015; Jahodářová et al., 2022) for detailed identification of genetic CRISPR elements in depth. Complementary functional annotation and phylogenetic classification of the genome-encoded enzymes according to the Clusters of Orthologous Groups of proteins (COGs) were performed using eggNOG-mapper (Emms & Kelly, 2018; Kelly & Maini, 2013). The full-length ribosomal 16S rRNA gene sequence of the investigated strain, as extracted from the genome with barrnap (Seemann, 2013), showed highest similarities with an uncultivated cyanobacterium from the Sagole hotspring (Jarett et al., 2014), South Africa, with an identity of 99.6% and P. lividus PCC 6715 with an identity of 98.78% and according to a blastn suite search, utilizing the rRNA/ITS databases as reference (Zhang et al., 2000).
Quality control and in silico analyses
Deploying the public Galaxy.eu server of the Galaxy web platform (Emms & Kelly, 2017), the genome assembly quality was assessed regarding completeness and contamination with checkM (Emms & Kelly, 2019). In that context, CheckM was run through the browser-based tool Protologger, which is also part of the Galaxy network (Letunic & Bork, 2024). Additionally, the genome completeness was analyzed based on orthologous genes with BUSCO 5.4.6 (Ritz et al., 2023), utilizing the cyanobacteria_odb10 lineage. Whole-genome alignments were performed using the progressiveMauve (Koren et al., 2017) plugin within the Geneious Prime 2022.0.1 software. The circular plot was created using CIRCOS in a Linux Ubuntu environment (Li et al., 2021). Biosynthetic gene clusters were identified utilizing antiSMASH 7.0 (Tatusova et al., 2016).
2.4 Proteomics
2.4.1 Sample preparation
For the proteome analysis, 50 mL of cyanobacterial culture was harvested and centrifuged at 5000g for 10 min. Afterward, the biomass pellet was frozen with liquid nitrogen and stored at −80°C. Proteome analysis was performed in triplicates following a previously described protocol (Haft et al., 2018). For sample preparation, the pellets were resuspended in 600 µL in aqueous solution with 2% SDS and 10 mM β-mercaptoethanol. Next, the samples were incubated at 65°C for 5 min and centrifuged at 14,000 rcf for 10 min. The pellet was resuspended in 600 µL of methanol and vortexed. Then, 150 µL of chloroform was added and the sample was vortexed again. At last, 450 µL of dH2O was added. After vortexing, the samples were centrifuged at 14,000 rcf for 1 min. This step resulted in three separate phases. The middle phase, containing the proteins, was carefully isolated and mixed with 400 µL of methanol. Afterward, the samples were centrifuged at 14,000g for 2 min. After removing the supernatant, this step was repeated twice. Then, the samples were mixed with 400 µL of acetone, centrifuged at 14,000 rcf for 2 min, and the supernatant was discarded. This step was also repeated twice. The pellet was dried at room temperature for 25 min. Then, the pellet was resuspended in an aqueous solution with 8 M urea and 10 mM β-mercaptoethanol and stored at −20°C.
Proteins were run 1 cm into a 10% Criterion™ Tris–HCl Protein Gel (Bio-Rad Laboratories Inc.) and stained with Coomassie Brilliant Blue (SERVA Electrophoresis GmbH). Then, protein bands were excised from the gel and subsequently used for peptide isolation. The excised gel pieces were cut into small pieces (<1 mm3) and washed with acetonitrile until the Coomassie Brilliant Blue was fully removed. Afterward, the gel pieces were dried under vacuum for 15 min (GeneVac Evaporator; GeneVac HiTechTrader).
The dried samples were resuspended in 100 µL 10 mM Dithiothreitol solution and incubated for 45 min at 56°C in a shaker at 550 rpm. Then, the samples were washed with acetonitrile and incubated in 100 µL 55 mM iodoacetamide and 50 mM ammonium bicarbonate for 30 min in the dark in a shaker at 550 rpm. Afterward, the samples were washed twice with acetonitrile and dried under vacuum for 15 min (GeneVac Evaporator; GeneVac HiTechTrader). Then, samples were resuspended in 100 µL of digest solution containing 1 µL trypsin solution in 100 µL 25 mM ammonium bicarbonate buffer. The enzymatic digestion was performed overnight at 37°C in a thermal shaker at 300 rpm. Afterward, the peptides were extracted by adding 25 mM ammonium bicarbonate and a subsequent sonification for 15 min. After the addition of 100 µL acetonitrile and incubation for 15 min, 100 µL 5% formic acid was added, followed by a second sonification for 15 min. Then, 100 µL acetonitrile was added and the supernatant was transferred to a new microreaction tube. This step was repeated, and the samples were dried under vacuum (GeneVac Evaporator; GeneVac HiTechTrader). The dried peptide samples were mixed with 20 µL 1% formic acid, sonicated for 10 min, and filtered through a 10 kDa spin filter. Subsequently, the samples were subjected to LC-MS/MS analysis.
2.4.2 LC-MS/MS analysis
Peptide analysis was done using an LC-MS/MS analysis with a timsTOF Pro mass spectrometer equipped with a NanoElute LC System (Bruker Daltonik GmbH) on an Aurora column (250 × 0.075 mm, 1.6 µm; IonOpticks). The mobile phase comprised a 0.1% (v/v) water–formic acid mixture (A) and a 0.1% (v/v) acetonitrile–formic acid mixture (B), which was added as a binary gradient at a flow rate of 0.4 μL/min. The gradient concentration started at 2% (v/v) B and was increased linearly to 17% B after 36 min. After another 18 min, the percentage of C was increased to 25% (v/v) and then increased linearly to 37% B after 6 min. After 70 min, the concentration of B was adjusted to the final value of 95% (v/v). The oven temperature during the measurements was 50°C.
The timsTOF Pro mass spectrometer (TIMS) was used in PASEF mode with the following settings: mass range: 100–1700 mass: charge [m/z] ratio; ion mobility ramp, 0.6–1.6 V s/cm2; 10 MS/MS scans per ion mobility ramp (total cycle time 1.16 s); charge range, 0–5; active exclusion for 0.4 min; a target intensity of 20,000 counts; and an intensity threshold of 1000 counts. The collision energy was ramped stepwise, appropriate to the ion-mobility ramp, from 20 to 59 eV. The electrospray ionization source parameters were 1600 V for the capillary voltage and 3 L/min N2 (as dry gas) at a dry gas temperature of 180°C. The measurements were performed in a positive ion mode. Mass calibration was performed using the sodium formate cluster, and the TIMS was calibrated using Hexais (2,2-difluoroethoxy) phosphazene, Hexakis (2,2,3,3-tetrafluoropropoxy) phosphazene, and Chip cube high mass references (m/z ratios of 622, 922, and 1222, respectively).
2.4.3 Bioinformatic analyses
The MS/MS tandem spectrometry data of tryptic-digested peptides were evaluated using the PEAKS studio software (v.10.6, build 20201221). The annotated genome of Thermosynechococcaceae cyanobacterium sp. Okahandja (this study) (BioSample accession SAMN37714564) served as a reference for protein identification (Ciufo et al., 2018). The database search parameters included a precursor mass of 25 ppm, employing monoisotopic mass, and a fragment ion error tolerance of 0.05 Da. Trypsin was used as the digestion enzyme, allowing a maximum of two missed cleavages per peptide. Up to three variable PTMs were allowed per peptide, and FDR estimation was performed using decoy fusion. For protein identification, a 1.0% FDR threshold was applied, requiring at least one unique peptide per protein. The included PEAKSQ software was used to differentially quantify the three temperature sample groups, consisting of technical triplicates, each. A mass error tolerance of 20.0 ppm, an ion mobility tolerance of 0.05 Da, and a retention time shift tolerance of 6 min (auto detect) were employed. Fold change and significance thresholds were set to 2 and 15, respectively, utilizing the analysis of variance significance method. All proteins were exported for manual evaluation and plot creation. RStudio (Lomsadze et al., 2018) with the ggplot2 package (Haft, 2001) and Inkscape 1.2.2 were utilized to create all heatmaps, bar charts, and plots unless stated otherwise.
3 RESULTS AND DISCUSSION
3.1 Morphology
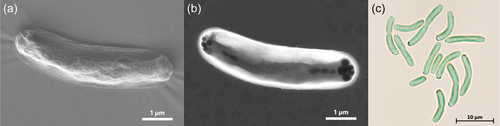
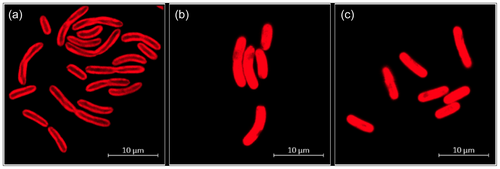
The cyanobacteria cells had a cylindrical, usually feebly curved form with a diameter of 1–2 µm and a length of 4–9 µm (Figure 1c). Dividing cells were frequently connected as linear pairs. Dark granula at the poles of the cells (Figure 1b) might represent cell deposits for energy storage, such as glucans or polyhydroxybutyrate (PHB). The distribution of thylakoids within the cells differed depending on the temperature (Figure 2). At a cultivation temperature of 40°C, the thylakoids were predominantly located in the areas near the cell membrane. At 50°C and 55°C however, they appeared to be evenly distributed. Nevertheless, temperature differences did not affect the overall cell shape and size.
3.2 Whole-genome sequencing
Whole-genome sequencing with the PacBio Sequel IIe instrument yielded 859,882 >Q20 reads comprising over 6.4 billion base pairs, with a median read quality of Q36 and a mean >Q20 read length of 10,893 bp. The Canu v2.0 assembler (Tang et al., 2018) was fed with the raw read FASTQ files and recognized 49,509 reads with 540 million bases, equal to a 200-fold genome coverage. After trimming, roughly 108 million bases from 6242 reads were further processed, corresponding to a 40-fold genome coverage. One circular contig with a size of 2.8 Mbp could be constructed, similar in size to the P. lividus PCC 6715 reference genome size of 2.65 Mbp on NCBI (RefSeq NZ_CP018092.1).
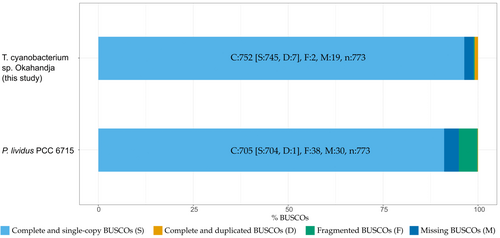
Genome quality was assessed with the tools checkM, according to which the genome completeness amounts to 99.76% with a contamination of 1.06% (Emms & Kelly, 2019), in addition to BUSCO v.5.4.6 which operates based on near-universal single-copy orthologs (Saxena et al., 2017). Comparing the gene set of the investigated Thermosynechococcaceae cyanobacterium sp. Okahandja genome to the cyanobacteria lineage data set (cyanobacteria_odb10) which consists of 141 genomes, a genome completeness of 97.3% was predicted (Figure 3). Interestingly, the reference genome of P. lividus PCC 6715 is estimated to possess merely 91.2% of all orthologous cyanobacterial genes, with a higher proportion of fragmented BUSCOs.
3.3 Phylogenetic analysis
3.3.1 16S rRNA gene sequence-inferred phylogeny
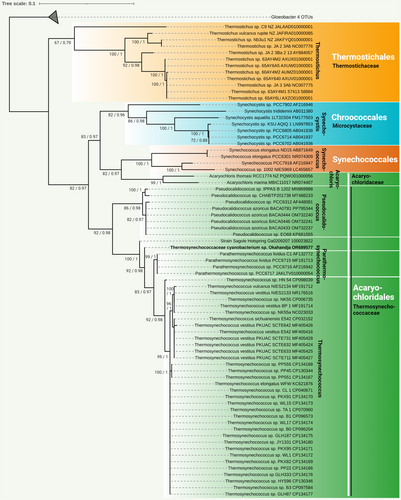
The complete genome of the newly isolated cyanobacterial strain was sequenced in this study. Based on the extracted 16S rRNA gene sequence, its phylogenetic position was analyzed (Figure 4). The cyanobacterial strain clustered well supported with the unicellular, thermophilic species P. lividus PCC 6715. These cyanobacteria are closely related to the genus Thermosynechococcus and display a more distant relationship with the genus Synechocystis and the species Synechococcus elongatus ND15, Synechococcus elongatus PCC 6301, Synechococcus sp. PCC 7918, and Synechococcus sp. 1002 NIES969 (Shih et al., 2013; Strunecký et al., 2023). Based on these findings and a comparison with known 16S rRNA gene sequences using the BLAST tool of GenBank (Huerta-Cepas et al., 2019), the isolated cyanobacterial strain was categorized as a novel species within the Thermosynechococcaceae family (Figure 4). Further phylogenetic analysis based on the 16S rRNA gene assigned the strain to a separate branch, closely related to the P. lividus strains PCC 6715-6717 (formerly classified as Thermosynechococcus lividus, Synechococcus lividus, or Thermostichus lividus) (Komárek et al., 2020).
3.3.2 Whole-genome sequence-inferred phylogeny
Average nucleotide identity (ANI) analysis of the investigated strain, an established whole-genome sequence-based method to identify prokaryotic species (Huerta-Cepas et al., 2017; Jarett et al., 2014), yielded maximum identities of approximately 81% with P. lividus PCC 6715. However, the threshold for reliable ANI-based species identification is 95% and above, with few exceptions, for example, Variovorax spp. with 88%, reflecting a less distinct species relation within the genus (Seemann, 2013). At the same time, progressiveMauve (Koren et al., 2017)-guided whole-genome alignment with the reference strain P. lividus PCC 6715 (Mammoth Hot Spring, Yellowstone National Park) revealed substantial genetic differences (Figure A1), with many gene inversion events and non-conserved regions, underpinning the role of geographical separation for gene drive. Furthermore, a digital DNA-DNA hybridization based on a Genome Blast Distance Phylogeny (GBDP) approach was deployed on the free GGDC (genome-to-genome distance calculator) web service of the German Strain collection (http://ggdc.dsmz.de) (Auch et al., 2010) between the genomes of the strain of this study and P. lividus PCC 6715, its closest relative (Meier-Kolthoff et al., 2013, 2022). In the process, three distinct formulas were applied to infer genome-to-genome distances based on obtained sets of HSPs (high-scoring segment pairs) and MUMs (maximal unique matches). These distances were transformed to values analogous to DDH using a generalized linear model (GLM) inferred from an empirical reference data set comprising real DDH values and genome sequences. Of the three formulas, two—including the recommended formula—concluded that the genome of our study represents a distinct genome compared with that of the strain PCC 6715. Furthermore, it concluded based on the GC%-difference of 1.53% that the two genomes belong to distinct species, since the %GC content cannot differentiate more than 1% (Meier-Kolthoff et al., 2014). Refer to the supplementary material for details on the evaluation with GGDC: https://zenodo.org/doi/10.5281/zenodo.10007199.
3.3.3 Orthology-inferred phylogeny
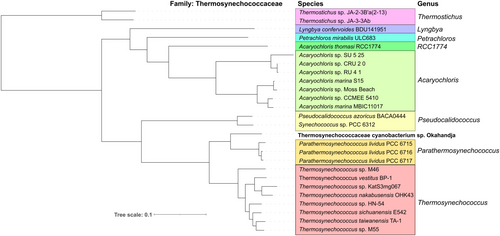
Investigation of representative Thermosynechococcaceae genomes for orthologous proteins yielded a total of 451 orthogroups (including gene duplication events) and 270 single-copy genes (exactly one gene per species) present in all genomes (Figure 5). When only comparing the eight delineated Thermosynechococcus species and the three P. lividus strains PCC 6715, PCC 6716, and PCC 6717 according to Tang et al. (2024) with the novel strain of this study, 1476 orthogroups encompassing 1268 single-copy genes were detected. When regarding the genomes of Parathermosynechococcus spp. and the novel strain from Okahandja exclusively, 2215 common orthogroups and 2090 single-copy genes were identified. Subsequent multiple sequence alignment and phylogenetic tree construction based on the ortholog-inferred phylogenetic tree assigned the Thermosynechococcaceae cyanobacterium sp. Okahandja of this study to a distinct and separate clade besides the Yellowstone National Park-derived genus of Parathermosynechococcus (Figure 5) (Tang et al., 2024). Therefore, the orthology-inferred phylogenetic analysis supports the results of the 16S rRNA gene sequence-inferred phylogenetic tree (Figure 4), suggesting the existence of a novel genus comprising two species so far (strain Sagole Hotspring and Thermosynechococcaceae cyanobacterium sp. Okahandja).
3.3.4 Morphological and phylogenetic characterization suggests a novel species belonging to the family of Thermosynechococcaceae
P. lividus PCC 6715 was formerly known as Thermosynechococcus lividus or Synechococcus lividus, belonging to the order of Synechococcales. Recently, it was proposed to belong to a new genus ‘Parathermosynechococcus’ based on GTDB classification, ANI/AAI, and phylogenomics by Tang et al. (2024). As discussed above, the newly isolated strain from this study was very closely related to the strain P. lividus PCC 6715 based on their 16S rRNA gene sequences. In that context, the morphological attributes of the cyanobacterial cells observed under the microscope (Figure 1) resembled previous descriptions in the literature for this species (Copeland, 1936; Tang et al., 2024). Taking into account that the investigated strain of this study was assigned to a separate branch than the strain P. lividus PCC 6715 based on 16S rRNA gene sequence homology and a whole-genome sequence-based aligned nucleotide identity (ANI) of only 81%, we propose to assign the strain of this study to the family Thermosynechococcaceae. Based on the position within the phylogenetic trees, there is an indication that this strain might belong to a novel genus. However, this hypothesis should be confirmed with more certainty if other closely related species are discovered in the future. Further data to support this view are differences in the genome-wide GC content of 55% (this study) as opposed to 53.5% of P. lividus PCC 6715 and the whole-genome sequence alignment with progressiveMauve (Koren et al., 2017) (Figures A1 and 7), which revealed dramatic genome fluctuation in the form of gene deletions, rearrangements, and inversion events. Papke et al. (2003) revealed the importance of geographic separation for allopatric speciation events in cyanobacteria using genetic marker investigation of bacterial hotspring populations worldwide (Parks et al., 2015). The authors emphasized that the hypothesis “everything is everywhere” is incorrect and that lateral gene transfer microbial community dynamics alone are not sufficient to describe speciation accurately. In accordance with many other reports (Blin et al., 2023; Darling et al., 2010; Fuchs et al., 2021; Gao et al., 2004; Hitch et al., 2021; Krzywinski et al., 2009; Manni et al., 2021; Ward et al., 2012), our data appears to confirm the thesis of Papke and colleagues.
3.3.5 Phylogenetic classification according to COG
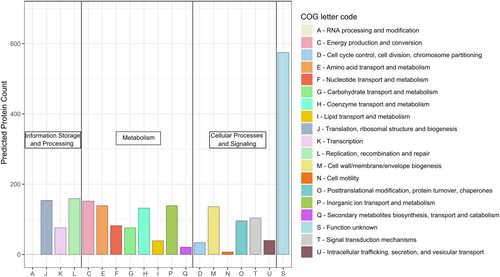
Phylogenetic classification of the genome-encoded proteins according to the Clusters of Orthologous Groups of proteins (COGs) revealed an annotation ratio of approximately 70%, with 30% proteins of unknown function (Figure 6). Like most cyanobacteria, Thermosynechococcaceae cyanobacterium sp. Okahandja features little to no proteins involved in RNA processing and modification (A), chromatin structure and dynamics (B), extracellular structures (W), nuclear structure (Y), or a cytoskeleton (Z) (Ma et al., 2003). The general COG distribution pattern is in accordance with that of other Thermosynechococcaceae cyanobacteria, with noticeably higher fractions of translation than transcription and amino acid metabolism-related proteins than carbohydrate metabolism-related proteins (Adomako et al., 2022).
3.4 In silico analyses and visualization of the novel genome
3.4.1 Whole-genome alignment reveals profound gene flux
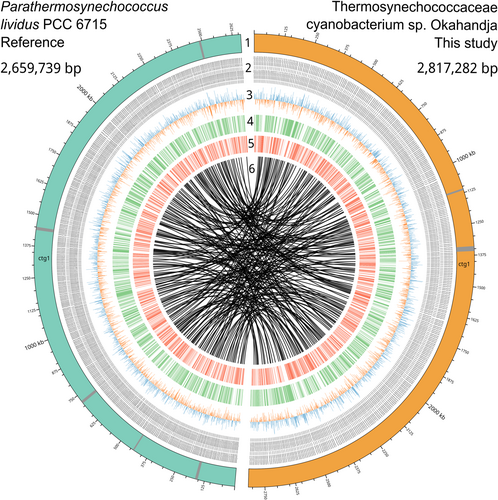
To investigate conserved region abundance and general genomic arrangement, a whole-genome alignment of the closest relative, P. lividus PCC 6715, and Thermosynechococcaceae cyanobacterium sp. Okahandja was performed using progressiveMauve (Figure A1). Deploying a locally collinear block (LCB) weight of 4000 bp, 212 conserved regions shared among the two species could be detected. As apparent by the multitude of overlapping LCB-connecting lines, the localization of individual conserved regions between the two aligned genomes diverges heavily. Furthermore, as indicated by the second, lower row in the depicted Thermosynechococcaceae cyanobacterium sp. Okahandja genome, about half of the conserved regions have been inverted in their respective orientation.
Finally, as evident through the numerous white gaps in between colored, LCB-representing boxes, many genes are neither shared nor conserved between P. lividus PCC 6715 and Thermosynechococcaceae cyanobacterium sp. Okahandja, underpinning their genomic disparities. Considering that the genomes differ by 150 kb in size, it is physically impossible for all genetic elements to be conserved independently of their localization.
3.4.2 Circular genome plot and phage region investigation
A comparative circular plot serves to illustrate genomic similarities and disparities between Thermosynechococcaceae cyanobacterium sp. Okahandja of this study and P. lividus PCC 6715 (Figure 7). Since both genomes were derived from PacBio long-read sequencing platforms, circular chromosomes could be resolved within one continuous contig via the assembly. PHASTER-guided prediction revealed two incomplete phage regions for Thermosynechococcaceae cyanobacterium sp. Okahandja and five incomplete phage regions for P. lividus PCC 6715 (Team, 2022), which is indicated by the gray bands in the respective chromosomes (colored teal or orange). Visualization of the GC/AT skews uncovered balanced nucleotide distributions throughout the whole cyanobacterial genomes, in contrast to those of many bacteria, where regions of GC or AT overabundance persist (Wickham, 2008). The innermost circle depicts the results from the progressiveMauve whole-genome alignment (Koren et al., 2017) (Figure A1), emphasizing the multitude of conserved regions (origin of lines), their genomic rearrangement (lines), and non-conserved regions (white gaps).
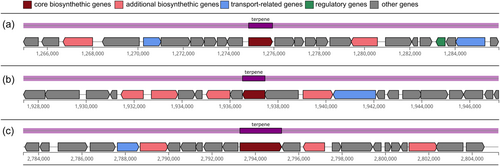
3.4.3 Thermosynechococcaceae cyanobacterium sp. Okahandja harbors three biosynthetic gene clusters for terpenes
Inquiry for industrially relevant secondary metabolite production genes with antiSMASH 7.0 (Blin et al., 2023) revealed three terpene-producing biosynthetic gene clusters (BCGs) of approximately 20 kbp length, harboring one core gene each (Figure 8). Hereby, the terpene synthesizing proteins of the BCGs I–III were annotated as 6-carboxytetrahydropterin synthase, phytoene synthase, and squalene-hopene cyclase, respectively. Opposed to many cyanobacteria that belong to more recent evolutionary clades, no BCGs for non-ribosomal peptide, bacteriocin, or polyketide production could be detected in our investigated strain (Ma et al., 2003). Phytoene is a colorless carotenoid intermediate, exhibiting an unusually low number of three conjugated double bonds and acting as a precursor for other carotenoids (Strunecký et al., 2023). Consequently, phytoene absorbs light at the UVB region (280–320 nm) and is structurally less rigid. Implications are potentially photoprotective features against UV-B light-induced skin cancers (Shih et al., 2013) and a significantly higher bioaccessibility (Strunecký et al., 2023), partly because aggregation and crystal formation are less likely (Camacho et al., 2009). Data also suggest that the colorless carotenoids play a more important role in nutrition than previously thought, with intake amounts exceeding those of the major dietary carotenoids lycopene, lutein, β-cryptoxanthin, violaxanthin, neoxanthin, and zeaxanthin (Komárek et al., 2020). Therefore, the phytoene synthase of Thermosynechococcaceae cyanobacterium sp. Okahandja might represent an interesting candidate for recombinant carotenoid production.
3.4.4 Insights from inspection of the CRISPR/Cas machinery
Functionally, the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) adaptive immune system acts as a bacterial defense mechanism to deflect invading mobile elements, including phage attacks (Westra et al., 2016). Jahodářová et al. examinated the CRISPR/Cas machinery evolution in Thermostichus spp. (Jahodářová et al., 2022), which uncovered differences in CRISPR spacer diversity based on geographically exclusive phage predation pressure. Spacer regions are phage genome-derived sequences incorporated by singular bacteria cells into their CRISPR arrays to acquire resistance against that specific bacteriophage (Barrangou et al., 2007), a phenotype that is inherited by future clones (Koonin & Wolf, 2009; Wang et al., 2016; Westra et al., 2016). Following that example, we inquired about the novel genome for CRISPR-associated genes, which are more abundant and longer among thermophiles (Anderson et al., 2011), revealing 19 genome-encoded proteins with eggNOG-mapper (Huerta-Cepas et al., 2019) and 18 with RAST (Aziz et al., 2008; Brettin et al., 2015; Overbeek et al., 2014), belonging to class 1 (Jahodářová et al., 2022; Westra et al., 2016). Contrarily, the closest relative, P. lividus PCC 6715, which originated from the Octopus Spring in the Yellowstone National Park (Dyer & Gafford, 1961), exhibited merely two CRISPR-associated genes over 2 arrays, encompassing 10 spacers and 12 repeats (Table 1). The second member of the genus Parathermosynechococcus (Tang et al., 2024), strain PCC 6717, which was also sampled in the Yellowstone National Park (Fairy Spring), exhibited similarly low CRISPR-associated gene numbers with 9 over 3 arrays, encompassing 22 spacers and 25 repeats. The third specimen, P. lividus PCC 6716, was derived from the Hunter's Hot Spring in Oregon and has a comparably extensive CRISPR/Cas system as the investigated strain from Namibia. Genome annotation with RAST (Aziz et al., 2008; Brettin et al., 2015; Overbeek et al., 2014) revealed a total of 19 CRISPR-associated genes, 69 spacers, and 74 repeats distributed over 5 arrays.
| Strain | Arrays | Genes | Repeats | Spacers | Origin | Year |
|---|---|---|---|---|---|---|
| PCC 6715 | 2 | 2 | 12 | 10 | Octopus Spring, Yellowstone National Park | 1961 |
| PCC 6716 OH-53s | 5 | 19 | 74 | 69 | Hunter's Spring, Oregon | 1962 |
| PCC 6717 Y52s | 3 | 9 | 25 | 22 | Fairy's Spring, Yellowstone National Park | 1964 |
| Okahandja | 6 | 18 | 137 | 131 | Hotspring near Okahandja, Namibia | 2024 |
CRISPR spacer sequence alignments of Thermosynechococcaceae cyanobacterium sp. Okahandja with P. lividus PCC 6715 revealed no shared spacer regions, confirming yet again that CRISPR arrays target local rather than global phages (Kunin et al., 2008) and that the antagonistic host–virus coevolution outpaces bacterial dispersal (Berg Miller et al., 2012; Westra et al., 2016). Furthermore, a nucleotide BLAST search of all CRISPR spacer sequences resulted in no matches with virus genome sequences, which is a common phenomenon (Anderson et al., 2011). This could be attributed to CRISPR/Cas targeting rare viruses only (Emerson et al., 2013) but more likely due to the limited accessibility of phage sequences (Andersson & Banfield, 2008; Westra et al., 2016).
Analogous to the thermophilic PCC 6715, the two Thermostichus strains JA2-3B'a (Abed et al., 2009; Burra et al., 2010; Fish & Codd, 1994; Galtier & Lobry, 1997; Graverholt & Eriksen, 2007; Kumar et al., 2019; Nishioka et al., 2001; Patel et al., 2019; Sabath et al., 2013; Thompson & Eisenberg, 1999; Van Noort et al., 2013; Ward et al., 2012) and 3-3Ab were derived from the Octopus Spring (Yellowstone National Park) (Allewalt et al., 2006). Interestingly, the analysis of Jahodářová et al. revealed far more extensive CRISPR/Cas machinery for the two Thermostichus spp. compared to PCC 6715 despite their identical habitat (Jahodářová et al., 2022). The extreme time discrepancy between the sample collection of PCC 6715 in 1961 (Dyer & Gafford, 1961) and the two Thermostichus specimens in 2006 may serve to explain the divergent phage predation pressures and provide evidence for the continuous coevolution.
The importance of the CRISPR immune response in phage attacks is further illustrated in the increased prophage sequence count (dormant, non-lytic phage sequences) in the genome of PCC 6715 (five, refer to Figure 7) as opposed to merely one prophage sequence in Thermosynechococcaceae cyanobacterium sp. Okahandja, the latter with a more complex CRISPR/Cas system at its disposal. A possible conclusion would be that the various horizontal gene transfer (HGT) events, visualized by the whole-genome sequence alignment with progressiveMauve (Darling et al., 2010), happened to PCC 6715 but not to the better-defended strain evaluated in this study, implying that Thermosynechococcaceae cyanobacterium sp. Okahandja is genetically closer to the common progenitor. It has been demonstrated that the CRISPR/Cas system can also act as an HGT barrier (Erdmann & Garrett, 2012; Palmer & Gilmore, 2010), although contradicting evidence has been reported (Gophna et al., 2015; Westra et al., 2016), as well. However, the focal point of this hypothesis is the time point of CRISPR/Cas system acquirement or loss thereof, depending on which of the two scenarios holds true.
3.5 Differential proteome analysis uncovers 129 heat-dependent proteins
Evaluation of the intracellular Thermosynechococcaceae cyanobacterium sp. Okahandja proteomes derived from cultivations conducted at 40°C, 50°C, or 55°C revealed 43 upregulated proteins at elevated temperatures (Figure 9). Overall, if one includes the proteins with reduced expression levels at elevated temperatures, 129 differentially expressed, thermosensitive proteins could be identified (supplementary material for the complete list: https://zenodo.org/doi/10.5281/zenodo.10007199). It is important to note that samples were normalized based on the 40°C controls; thus, all fold changes at 50°C or 55°C are depicted with respect to protein abundancies at the lowest temperature condition. To that end, seven proteins showed increased abundances at 50°C, exclusively, followed by decreased expression levels at 55°C (Gene IDs 3, 363, 418, 1061, 1993, 2070, 2711). Additionally, 17 proteins were 1.2-fold (or 20%) more abundant at 55°C, but not at 50°C, compared to the 40°C condition (gene IDs 93, 104, 153, 421, 477, 590, 672, 1055, 1171, 1406, 1483, 1516, 1611, 1715, 1860, 1933, and 2550). A singular protein (gene ID 1859) was upregulated at both elevated temperatures, with a maximum abundance at 50°C. The remaining 18 proteins (gene IDs 100, 859, 917, 950, 960, 1222, 1260, 1317, 1681, 1693, 1699, 1772, 1980, 2046, 2469, 2505, 2580, 2673) were increasingly upregulated with elevated temperature, reaching fold changes of up to 13 at 55°C.
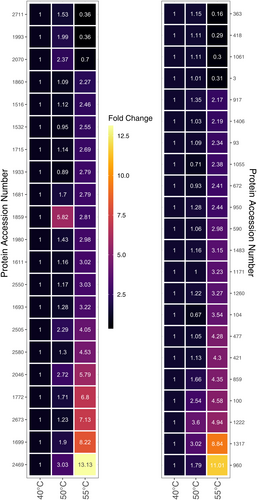
Detailed inspection of the 10 topmost upregulated enzymes at elevated temperatures (Table 2) uncovered three hypothetical proteins, two of which could be annotated as nuclease activity containing protein (gene ID 2469) and signal transduction histidine kinase (gene ID 960), respectively, with eggNOG-mapper 5.0 (Huerta-Cepas et al., 2019). Other proteins included a dTDP-4-dehydrorhamnose 3,5-epimerase (gene ID 1699), an ergothioneine biosynthesis protein EgtC (gene ID 1859), a Ycf51 family protein (gene ID 1222), a type II toxin–antitoxin system VapC family toxin (gene ID 1772), a crossover junction endodeoxyribonuclease RuvC (gene ID 2046), the 50S ribosomal protein L33 (gene ID 2673), and a carbohydrate ABC transporter permease (gene ID 100). Except for the ergothioneine biosynthesis protein EgtC, all proteins were more abundant at 55°C than at 50°C with fold changes between approximately 1.4-fold for the Ycf51 family protein and sixfold for the putative signal transduction histidine kinase.
| Gene ID | Log2 FC (50°C) | Log2 FC (55°C) | Annotation | ||
|---|---|---|---|---|---|
| PGAP | eggNOG | KEGG | |||
| 2469 | 3,03 | 13,13 | Hypothetical protein | Nuclease activity | No hits |
| 960 | 1,79 | 11,01 | Hypothetical protein | Signal transduction histidine kinase | No hits |
| 1317 | 3,02 | 8,84 | Hypothetical protein | No hits | No hits |
| 1699 | 1,9 | 8,22 | dTDP-4-dehydro-rhamnose 3,5-epimerase | Catalyzes the epimerization of the C3′ and C5′ positions of dTDP−6-deoxy-d-xylo-4-hexulose, forming dTDP-6-deoxy-l-lyxo-4- hexulose | rfbC, rmlC; dTDP-4-dehydro-rhamnose 3,5-epimerase |
| 1859 | 5,82 | 2,81 | Ergothioneine bio-synthesis protein EgtC | TIGR03442 family protein (ergothioneine biosynthesis protein EgtC) | No hits |
| 1222 | 3,6 | 4,94 | Ycf51 family protein | PFAM Protein of function (DUF2518) | No hits |
| 1772 | 1,71 | 6,8 | Type II toxin–antitoxin system VapC family toxin | Large family of predicted nucleotide-binding domains | No hits |
| 2046 | 2,72 | 5,79 | Crossover junction endodeoxyribonu-clease RuvC | Nuclease that resolves Holliday junction intermediates in genetic recombination. Cleaves the cruciform structure in supercoiled DNA by nicking to strands with the same polarity at sites symmetrically opposed at the junction in the homologous arms and leaves a 5′-terminal phosphate and a 3′-terminal hydroxyl group | ruvC; crossover junction endo-deoxy-ribonuclease RuvC |
| 2673 | 1,23 | 7,13 | 50S ribosomal protein L33 | Belongs to the bacterial ribosomal protein bL33 family | RP-L33, MRPL33, rpmG; large subunit ribosomal protein L33 |
| 100 | 2,54 | 4,58 | Carbohydrate ABC transporter permease | ABC-type sugar transport system, permease component | chiG; putative chitobiose transport system permease protein |
3.5.1 Adaptation of protein expression patterns to elevated temperatures
Please refer to Table A2 in the appendix for a curated list of all discussed proteins in the following sections.
Modulation of the genetic reproduction machinery
RNAs and proteins involved in transcription and translation possess higher GC contents in thermophiles under elevated temperatures (Basak et al., 2004), but high genome GC compositions alone are no indicators of thermophily or at least not universally applicable (Basak et al., 2004; Wang et al., 2015; Zeldovich et al., 2007). With 57.3%, the average GC content of all 41 genomic encoded transfer RNAs, detected with tRNAscan (Lowe & Eddy, 1997), was indeed slightly above the genome-wide average GC distribution of 55%.
Among the 129 differentially expressed thermosensitive proteins, 13 were associated with gene transcription or translation. Of these, three proteins with nuclease activity (gene IDs 2046, 2469 & 2574) were identified, where two proteins were incrementally upregulated (gene IDs 2046 & 2469), while the remaining nuclease was incrementally downregulated with increasing temperature. While protein 2469 is annotated as nuclease by eggNOG-mapper based on sequence homology, 2046 is the crossover junction endodeoxyribonuclease RuvC involved in homologous recombination. Interestingly, RuvC endonuclease-like domain containing CRISPR/Cas class 2 effectors have been identified, suggesting a potential involvement in the adaptive immune system of protein 2469 (Mapelli-Brahm & Meléndez-Martínez, 2021). The last nuclease of the identified triplet was the exoribonuclease D downregulated 2.7-2.9-fold with increasing temperature and functionally relevant for tRNA maturation (Zuo et al., 2005).
Additional incrementally upregulated proteins associated with genetic reproduction encompass the polymerase III subunit delta (gene ID 1611) responsible for the high speed and processivity of the polymerase III (Jeruzalmi et al., 2001), which is in turn involved in DNA replication, homologous recombination, and mismatch repair according to the KEGG classification (Kanehisa et al., 2016); the 50S ribosomal protein L33 (gene ID 2673), and the phenylalanyl-tRNA synthetase beta chain pheT (gene ID 421).
In contrast, the following five proteins were downregulated incrementally with increasing temperature: the segregation and condensation protein A, relevant for chromosomal partition during cell division (Mascarenhas et al., 2002; Soppa et al., 2002); the 30S ribosomal protein S12 (gene ID 158), involved in 30S and 16S rRNA stabilization (Demirci et al., 2013); the polyribonucleotide nucleotidyltransferase (gene ID 594), involved in mRNA degradation (Kleppe et al., 1971); the DNA-3-methyladenine glycosylase (gene ID 2513), responsible for base excision repair (Wyatt et al., 1999); and the DNA adenine methylase (gene ID 2324), realizing mismatch repair (Low et al., 2001). According to eggNOG-mapper (Huerta-Cepas et al., 2017; Huerta-Cepas et al., 2019), second putative chromosome segregation enforcing protein (gene ID 2711) was detected to be upregulated 1.5-fold at 50°C and then downregulated 2.8-fold at 55°C. In addition, a third ribosome-associated protein, the translation inhibitor RaiA (gene ID 2297), was steadily downregulated approximately threefold at both 50°C and 55°C. It is required for dimerization prevention of 70S ribosomes in the stationary phase (Ueta et al., 2005), reduces translation errors (Ueta et al., 2005), and inactivates 70S ribosomes during environmental stress in a reversible manner (Lang et al., 2021). Finally, a pentapeptide repeat-containing protein (gene ID 418) of unknown function, predicted to bind single-stranded DNA (Huerta-Cepas et al., 2017; Huerta-Cepas et al., 2019), was downregulated −3.5-fold at 55°C, exclusively.
While ribosomal proteins have been reported to be more abundant at high temperatures before, for example, in Bacillus methanolicus MGA3 and Pyrococcus furiosus (Müller et al., 2014; Trauger et al., 2008), the upregulation of the 50S ribosomal protein L33 is curious, as it does not have any significant role in ribosome synthesis or assembly in E. coli (Maguire et al., 1997). It could be hypothesized that either (i) L33 does play a pivotal role in the ribosome synthesis and assembly of Thermosynechococcaceae cyanobacterium sp. Okahandja acts as a physical “heat shield” for other critical cell parts or (ii) it is a side effect of the whole ribosomal protein operon being transcribed. Moreover, ribosomal protein complexes in thermophiles are reported to have stronger binding to 23S rRNA and more compact structures compared to those in mesophiles (Shcherbakov et al., 2006), which might secure the biosynthesis of functional proteins under adverse temperature conditions (Wang et al., 2015).
3.5.2 Differential expression of regulatory and CRISPR/Cas proteins
Closer inspection of regulatory proteins revealed a total of 11 proteins with expression levels depending on temperature conditions. Nine of these regulators were downregulated, whereas only two regulatory proteins were upregulated, namely a two-component system belonging to protein pixH required for phototactic motility in Synechocystis (Yoshihara & Ikeuchi, 2004), which was upregulated threefold (gene ID 1693) at 55°C, and a signal transduction histidine kinase (gene ID 960), which was upregulated 16-fold at the highest temperature. This histidine kinase might act as a global heat response regulator molecule, similar to σ32 in E. coli (Browning & Busby, 2004), paired with the 4.5-fold downregulated heat-inducible transcriptional repressor HrcA (gene ID 2557) controlling the transcription of class I heat shock genes (grpE- dnaK-dnaJ and groELS operons). Protein expression and characterization or knock-out experiments would have to confirm or deny the role of the histidine kinase in the thermal response.
AYcII family protein (gene ID 652) found to be fused to the RNA polymerase sigma-70 factor family domain and therefore possibly involved in transcription initiation (Yeats et al., 2003) and a YbaB/EbfC family nucleoid-associated protein (gene ID 1449) were downregulated the most, with 25- and 16-fold at 55°C, respectively. According to eggNOG-mapper (Huerta-Cepas et al., 2017; Huerta-Cepas et al., 2019), the latter binds to DNA and alters its conformation and may additionally be involved in the regulation of gene expression, nucleoid organization, and DNA protection. Interestingly, the third most downregulated protein at 55°C with eightfold was an ATP-binding histidine kinase (gene ID 302), annotated as phosphate regulon sensor histidine kinase PhoR by KEGG (Kanehisa et al., 2016). Other incrementally less abundant proteins with increasing temperature encompassed a GGDEF domain-containing protein (gene ID 1885), acting as diguanylate cyclase and cyclic di-GMP synthase (Paul et al., 2004; Ryjenkov et al., 2005); an ArsR/SmtB family metalloregulator (gene ID 1377), a LysR family transcriptional regulator (gene ID 1647); and the circadian clock protein KaiA (gene ID 1886). Finally, a translation elongation factor 4 protein (gene ID 2313), annotated as GTP-binding protein LepA, required for accurate and efficient protein synthesis under certain stress conditions, was downregulated approximately 3.8-fold at 50°C and 1.7-fold at 55°C.
Three temperature-dependent CRISPR/Cas proteins could be detected in the proteomic data set. However, no clear trend for a definitive up- or downregulation of the adaptive immune response proteins could be identified. To elucidate, an endonuclease Cas1 (gene ID 1513) was incrementally downregulated with increasing temperature, while a type I-E CRISPR-associated protein Cas7/Cse4/CasC (gene ID 1993) was upregulated at 50°C and downregulated at 55°C and finally a type III-B CRISPR-associated protein Cas10/Cmr2 (gene ID 1516), which was incrementally upregulated with rising temperature.
Potential role of a toxin–antitoxin system for thermal resistance
Bacterial type II toxin–antitoxin systems, a component of which was upregulated almost sevenfold at 55°C, have been reported before to convey heat resistance, for example, the ParDE system in Enterococcaceae (Kamruzzaman & Iredell, 2019) or in the hyperthermophilic crenarchaeon Sulfolobus solfataricus (Cooper et al. 2009). Toxin–antitoxin operons are a prevalent genetic feature found in bacteria (Pandey & Gerdes, 2005). These operons consist of two distinct genes: one encoding a harmful toxin and the other encoding a fragile antidote molecule (Kamruzzaman et al., 2021). In general, when transcription and translation processes are active, these genes produce proteins that interact to form a harmless complex. However, if the transcription or translation of the toxin–antitoxin operon is interrupted, the unstable antitoxin protein undergoes degradation through various mechanisms. This degradation leaves the toxin unchecked, allowing it to exert its biological impact (Overbeek et al., 2014). This impact can manifest as either killing the bacteria (bactericidal), inhibiting bacterial growth (bacteriostatic), or inducing a dormant metabolic state (Magnuson, 2007; Tsilibaris et al., 2007). The biological role and significance of bacterial toxin–antitoxin operons have sparked a wide array of hypotheses. These range from considering them as simple self-interested genetic elements disseminated through horizontal gene transfer to functioning as regulators of metabolic processes within the bacterial cell (Kamruzzaman et al., 2021; Magnuson, 2007; Tsilibaris et al., 2007).
In that regard, protein modeling with SwissModel (Studer et al., 2020; Waterhouse et al., 2018) yielded the highest score for a hypothetical protein PAE0151 of the hyperthermophilic archeon Pyrobaculum aerophilum localized in a PIN-domain toxin–antitoxin operon (vapBC operon) (Bunker et al., 2008). Additionally, the authors could demonstrate that PAE0151 (putative antitoxins) and adjacent PIN-domain proteins (putative toxins) are conserved in many thermophilic species, indicating their involvement in the heat response (Bunker et al., 2008).
Expression of photosynthesis-related proteins is attenuated during elevated temperatures
Investigation of the heat-dependent differentially expressed photosynthesis-related proteins suggests a strong trend toward downregulation at elevated temperatures. The data specifically indicated that of the nine detected proteins, all but one exhibited incremental downregulation with increasing temperature. In this context, it is noteworthy that the phycobilisome rod-core linker polypeptide (gene ID 1483) stood as a unique exception (Glauser et al., 1992). This photosynthesis-related protein displayed incremental upregulation, with fold changes of 1.15 and 3.15 at 50°C and 55°C, respectively. All other proteins, including the photosystem I reaction center subunit II (gene ID 694), the chlorophyll a-b binding domain-containing protein (gene ID 1748), the photosystem II manganese-stabilizing polypeptide (gene ID 1572), the photosynthetic/respiratory NAD(P)H-quinone oxidoreductase subunit C (gene ID 1413), the apocytochrome f (gene ID 2629), the phycocyanobilin lyase subunit beta (gene ID 1482), the protoporphyrinogen oxidase (gene ID 623), and the chlororespiratory reduction protein 7 (gene ID 697) were downregulated between 2.2- and ninefold at 55°C, respectively. Hence, attenuation of photosynthesis was corroborated by the proteomic data set.
Differential expression of transport, ion homeostasis, and redox-related proteins
Examining transport-related proteins in dependence of temperature revealed incremental downregulation of a cobalt/nickel importer (gene ID 2077), a lipopolysaccharide exporter (gene ID 506), a cysU, sulfate/thiosulfate transport system permease protein (gene ID 1624), the protein translocase subunit secF (gene ID 710), and a multicomponent Na + /H+ antiporter subunit B (gene ID 908). Contrarily, a SLC13 permease (gene ID 1681), annotated as Na + /H+ antiporter NhaD/arsenite permease-like protein by eggNOG-mapper (Huerta-Cepas et al., 2017; Huerta-Cepas et al., 2019), and a chitobiose import protein chiG (gene ID 100) were found to be incrementally upregulated with rising temperature, 3.8- and 4.6-fold at 55°C, respectively. Finally, the signal protein processing leader peptidase SppA (gene ID 1171) was upregulated threefold at 55°C, exclusively.
A closer inspection of ion homeostasis revealed a complex heat response expression pattern, with six protein abundancies correlating with temperature changes. Three proteins, among them a ferrous iron transport protein A (gene ID 1055), were upregulated at 55°C, exclusively, with the ferrous ion transport protein slightly downregulated at 50°C. The other two proteins were a SUMF1/EgtB/PvdO family nonheme iron enzyme (gene ID 1860), annotated as formylglycine-generating sulfatase enzyme by eggNOG-mapper (Huerta-Cepas et al., 2017; Huerta-Cepas et al., 2019), and a 2Fe–2S iron-sulfur cluster-binding protein (gene ID 93).
In contrast, two proteins, including a 2Fe–2S iron-sulfur cluster-binding protein (gene ID 2641) and a hypothetical protein (gene ID 1190), both annotated as ferredoxin by eggNOG-mapper (Huerta-Cepas et al., 2017; Huerta-Cepas et al., 2019), were downregulated incrementally with increasing temperature, 6.7-fold and threefold, respectively, at 55°C. Finally, a ferredoxin thioredoxin reductase variable alpha chain (gene ID 2070) was found to be upregulated approximately 2.7-fold at 50°C but downregulated 1.4-fold at 55°C.
Additionally, several redox protein levels were affected by heat, comprising the NADH-quinone oxidoreductase subunit J (gene ID 363) required for the respiratory chain (Verkhovsky & Bogachev, 2010), which was marginally upregulated at 50°C but downregulated sixfold at 55°C. Furthermore, a BCP, PRXQ, DOT5 thioredoxin-dependent peroxiredoxin, which catalyzes the reduction of H2O2 and organic hydroperoxides (Nelson et al., 2011; Reeves et al., 2011), and an AarF/ABC1/UbiB kinase family protein (gene ID 553), associated with ubiquinone biosynthesis in E. coli (Poon et al., 2000) or 2′-N-acetyltransferase in Providencia stuartii (Macinga et al., 1998), could be detected. Both enzymes were incrementally downregulated with increasing temperature, with fold changes of −7.7 and −4.4, respectively, at 55°C. Last but not least, a peptide methionine sulfoxide reductase MsrA (gene ID 2402), involved in the reversible oxidation–reduction of methionine sulfoxide in proteins to methionine and oxidative damage repair of proteins (Weissbach et al., 2002), was increasingly downregulated 1.35- and 4.6-fold at 50°C and 55°C, respectively. Consequently, Thermosynechococcaceae cyanobacterium sp. Okahandja seems to be more susceptible to oxidative stress with diminished oxidative respiration capabilities in exchange for increased fitness at elevated temperatures.
Metabolic adaptations to heat stress
When inspecting general metabolic proteins, we could identify 15 proteins with varying abundance depending on temperature (Table A2). Epitomizing the heavy metabolic burden on Thermosynechococcaceae cyanobacterium sp. Okahandja, 13 of these proteins were incrementally less biosynthesized at 50°C and subsequently 55°C normalized to expression levels at 40°C. While a GNAT family N-acetyltransferase (gene ID 104) transferring the acetyl residue from acetyl-coenzyme A onto diverse substrates (Shirmast et al., 2021) was upregulated 3.5-fold at 55°C and downregulated 1.5-fold at 50°C, solely a fructosamine kinase (Gemayel et al., 2007) (gene ID 917) was upregulated incrementally with rising temperature, 1.35 and 2.2-fold, respectively. The opposite was true for all other identified metabolic proteins, including the isoleucine-generating citramalate synthase (gene ID 955) (Howell et al., 1999); the glucose-6-isomerase (gene ID 2040), catalyzing the reversible isomerization of glucose-6-phosphate to fructose-6-phosphate (Hansen et al., 2001); and the chorismate mutase (gene ID 1170), which generates prephenate from chorismate in the shikimate pathway of tyrosine and phenylalanine biosynthesis (Hur & Bruice et al., 2003).
Interestingly, the most downregulated protein with 25-fold at 55°C is a squalene-hopene/tetraprenyl-beta-curcumene cyclase shc (gene ID 2698) according to KEGG (Kanehisa et al., 2016), which was also identified during our biosynthetic gene cluster analysis. The protein shows sequence similarity with a squalene-hopene cyclase (Hoshino et al., 2004; Sato et al., 2004) and a bifunctional triterpene/sesquiterpene cyclase (Sato et al., 2011a, 2011b), both products of which are not linked to heat resistance.
The second and third most downregulated proteins were a TldD/PmbA family protein (gene ID 1725), functioning as peptidase or N-deacetylase (Vobruba, 2022), which was downregulated first 6-fold and then 14-fold with increasing temperature, and the essential enzyme phosphoribosylglycinamide formyltransferase (gene ID 2289). It is required for the third step in purine biosynthesis (Aimi et al., 1990; Mullen & Jennings, 1996) and was downregulated approximately twofold at 50°C and 11-fold at 55°C. Although Thermosynechococcaceae cyanobacterium sp. Okahandja grew comparably well at 55°C compared to 50°C in our experiments, a long-term shortage of purines would drastically hamper the cell's genetic replication machinery. Even more so given their high abundance in mRNA of thermophilic organisms (Paz et al., 2004).
Further incrementally downregulated proteins with temperature increase included an M23 family metallopeptidase (gene ID 190), targeting peptidoglycans of the cell wall (Razew et al., 2022); two glycosyltransferase family 1 proteins (gene ID 2427 & 2708), transferring glycosyl moieties from activated sugar molecules to diverse acceptors (Zhang et al., 2020); a haloacid dehalogenase superfamily, subfamily IA (gene ID 1135); a deoxyhypusine synthase (gene ID 2533), required for homospermidine biosynthesis and biazotrophic growth in Anabaena spp. (Burnat et al., 2018); and a PFAM Glyoxalase bleomycin resistance protein dioxygenase (gene ID 2677), which is possibly involved in the antioxidant defense system of cyanobacteria, mediating detoxification of the abiotic stress response intermediate methylglyoxal (Rai et al., 2021; Suttisansanee & Honek, 2011).
Additionally, a NUDIX (nucleoside diphosphates linked to x) hydrolase (gene ID 2479), with the ability to hydrolyze a wide range of pyrophosphates (Bessman et al., 1996; McLennan, 2006), was detected to be fourfold less abundant at 55°C. While some Nudix superfamily members, such as MutT from E. coli, possess the ability to hydrolyze oxidized and potentially mutagenic nucleotides (McLennan, 2006), functions can also revolve around the control of metabolic intermediate or signaling compound levels (Bessman et al., 1996).
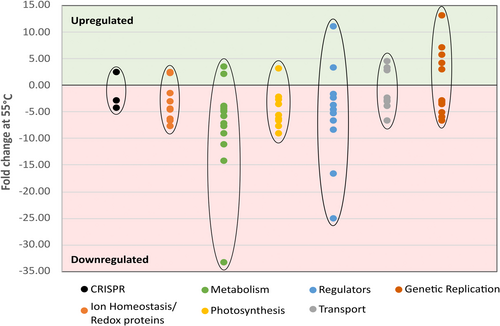
Finally, a bifunctional pantoate-beta-alanine ligase/dCMP kinase (gene ID 312) was detected as approximately 3.9-fold downregulated at 55°C throughout all samples. It represents a fusion of two enzymes that is unique to cyanobacteria according to the InterPro database (Blum et al., 2021; InterPro Entry IPR024894, 2024; Paysan-Lafosse et al., 2023). First, the pantoate-beta-alanine ligase catalyzes the condensation of pantoate with beta-alanine in an ATP-dependent reaction via a pantoyl-adenylate intermediate (Miyatake et al., 1979). Second, the dCMP kinase catalyzes the transfer of a phosphate group from ATP to either CMP or dCMP to form CDP or dCDP and ADP, respectively (Briozzo et al., 1998). Overall, apart from the phosphoribosylglycinamide formyltransferase, seemingly non-essential pathways were downregulated, hazarding the consequences of a higher vulnerability toward radical oxygen species and less metabolic flexibility. Figure 10 provides an overview of the fold changes at 55°C in relation to protein expression levels at 40°C of all discussed proteins. Refer to Table A2 for a comprehensive list of these proteins or the supplementary material for all 129 differentially expressed proteins in table form: https://zenodo.org/doi/10.5281/zenodo.10007199.
Bridging the gap: Integrating the findings with established research
Expectantly, in most thermophiles' proteomes such as Pyrococcus furiosus (Shockley et al., 2003), Thermotoga maritima (Pysz et al., 2004), or Thermoanaerobacter tengcongensis (Wang et al., 2007), heat shock proteins (HSPs) were abundant at elevated temperatures, forming complexes with other proteins to transmit thermal protection.
Despite the heat-inducible repressive HSP regulator HrcA (gene ID 2557) being strongly downregulated in the proteome of Thermosynechococcaceae cyanobacterium sp. Okahandja at 50°C and 55°C, thus theoretically enabling HSP upregulation, no actual heat shock protein fragments could be detected through MS/MS chromatography. Possible reasons for the lack of HSPs could be inadequate methodological sensitivity, incomplete protein extraction from cyanobacterial cells, or insufficiently high temperature during cultivation necessary for HSP induction in the investigated strain.
Consistent with the results from Chen et al. regarding the heat adaptations from T. tengcongensis (Chen et al., 2013), redox respiratory proteins such as ferredoxin, NADH-quinone oxidoreductase (subunit J), and NAD(P)H-quinone oxidoreductase (subunit C) were downregulated. Additionally, we found hints of (attempted) increased glucosamine recruitment through 4.5-fold upregulation of the putative chitobiose permease, which would be in line with the observed shift toward carbohydrate utilization for augmentation of energy generation in T. tengcongensis (Chen et al., 2013; Wang et al., 2007).
However, we could gather no evidence to support glucosamine to fructose conversion, as seen in T. tengcongensis (Chen et al., 2013). In that context, the data did generally also not suggest an increased metabolic flow toward fructose through enhanced expression of phosphomannomutase or glucosamine-6-P deaminase. Quite the opposite, the fructose-6-P generating glucose-6-isomerase (gene ID 2040) was downregulated incrementally with increasing temperature. However, continuous upregulation of phosphorylation catalyzing fructosamine kinase (gene ID 917) might indicate metabolic flow toward fructosamines formed by glycation. Although the importance of glycolysis in the heat response was demonstrated in systems biology-informed studies of several thermophiles including Thermotoga maritima (Wang et al., 2012), Geobacillus thermoglucosidasius (Loftie-Eaton et al., 2013), or Thermus thermophilus (Trauger et al., 2008), no glycolysis-related peptide fragments could be retrieved from our data set. Moreover, no further overlaps regarding upregulated proteins in response to elevated temperatures between Thermosynechococcaceae cyanobacterium sp. Okahandja and T. tengcongensis could be identified.
From the persistent downregulation of a chorismate mutase, also known as chorismate pyruvatemutase (gene ID 1170), and a citramalate synthase (gene ID 955), a potential lack of pyruvate could be deduced, as it is a substrate of both enzymes. In contrast to this observation, Chen et al. (2013) reported an increased flux toward pyruvate, mediated by glyceraldehyde-3-phosphate dehydrogenase. Furthermore, cistronic operon organization or gene clustering was reported in several other thermophilic organisms (Chen et al., 2013; Wang et al., 2015) but was not observed in our data set, where the differentially expressed proteins were rather scattered within the genome.
4 CONCLUSIONS
In this study, we present a novel thermophilic cyanobacterium species obtained from a hot spring near Okahandja, Namibia. PacBio-guided whole-genome sequencing of its genome and subsequent alignment with the closely related genome of P. lividus PCC 6715 revealed substantial gene flux events, putatively owed to local microbial community dynamics and ecological adaptations (Cadillo-Quiroz et al., 2012; Miller & Carvey, 2019; Schmutzer & Barraclough, 2019). Morphologically, it resembles other Thermosynechococcus spp. with its solitary, cylindrical cells and polar granulates, as determined with STEM, LEI, and brightfield microscopy (Komárek et al., 2020). Functional annotation with BlastKOALA (KEGG Orthology And Links Annotation) confirmed the two required genes for glycogen biosynthesis, glucose-1-phosphate adenylyltransferase, and starch synthase in the genome, as known from other cyanobacteria (Suzuki et al., 2013). While the 16S ribosomal RNA gene sequence exhibited a sequence similarity of 98.87% with that of the strain P. lividus PCC 6715, a maximum likelihood-based phylogenetic analysis (Figure 4) placed the isolate in a closely related, yet separate branch. Further indications for allopatric (geographically isolated) speciation were provided by a whole-genome sequence-based ANI of 81% between the two strains, with 95% representing the threshold for identical species (Ciufo et al., 2018).
Following the example of Jahodářová et al. (2022), we analyzed the CRISPR/Cas adaptive immune system and conducted ortholog-inferred phylogenetics of our isolated strain with respect to Parathermosynechococcus spp. and Thermosynechococcaceae, respectively. While insights about the local phage predation pressures of the respective strains could be gained (Westra et al., 2016), the vast difference in sampling time difference renders it difficult to compare their data. As expected, considering the geological and temporal distance between the strain of this study (Okahandja, Namibia, 2024) and P. lividus PCC 6715 (Yellowstone National Park, USA, 1961), no shared CRISPR spacer sequences could be detected. Furthermore, probably owed to the limited accessibility of phage sequences (Andersson & Banfield, 2008; Westra et al., 2016), no matches with virus genome sequences were yielded with a nucleotide BLAST search of all CRISPR spacer sequences.
In addition to the phylogenetic analysis based on 16S rRNA gene sequences, the orthology-based phylogenetic analysis (Figure 5) supported the notion that Thermosynechococcaceae cyanobacterium sp. Okahandja represents a novel genus within the family of Thermosynechococcaceae, which is closely related to the novel genus Parathermosynechococcus (Tang et al., 2024). Of its 2276 orthogroups, representing 93.8% of all genome-encoded genes, the Namibian strain shared 2257 orthogroups with P. lividus PCC 6715, while differing in 78 genes or 2.9% of its genome.
Furthermore, the present study shed light on the proteomic adaptations to varying temperatures of the isolated cyanobacterium. By investigating the overexpression of several key proteins in response to elevated temperatures, we have gained valuable insights into the mechanisms underlying the thermotolerance of this cyanobacterium. One important area of exploration is the identification and characterization of the specific genes and regulatory mechanisms that govern the observed overexpression of specific proteins in response to elevated temperatures. Understanding the molecular basis of these adaptations will provide valuable knowledge about the strategies employed by Thermosynechococcaceae cyanobacterium sp. Okahandja and other thermophilic cyanobacteria to thrive in extreme thermal environments. Genetic engineering approaches, such as gene knockout or overexpression, may offer opportunities to enhance the thermotolerance of other organisms or even develop novel biotechnological applications. An especially interesting candidate for such studies could be the phytoene synthase that was revealed through biosynthetic gene cluster inquiry.
Additionally, it would be intriguing to explore the broader ecological significance of the observed proteomic adaptations of Thermosynechococcaceae cyanobacterium sp. Okahandja. Understanding how these adaptations affect the overall fitness and ecological success of this cyanobacterium in its natural habitat, such as hot springs, would provide a more comprehensive picture of its role in these ecosystems. In this study, the adaptation of protein expression patterns of a new Thermosynechococcaceae cyanobacterium species to varying temperatures has provided valuable insights into the mechanisms underlying its thermotolerance. This research sets the stage for future investigations aimed at deciphering the regulatory pathways for the attainment of thermotolerance in cyanobacteria.
AUTHOR CONTRIBUTIONS
Nathanael D. Arnold: Conceptualization; methodology; software; validation; formal analysis; data curation; writing—original draft; writing—review and editing; visualization. Michael Paper: Conceptualization; methodology; software; validation; formal analysis; investigation; data curation; writing—original draft; writing—review and editing; visualization. Tobias Fuchs: Investigation. Nadim Ahmad: Investigation. Patrick Jung: Software; validation; visualization. Michael Lakatos: Resources; validation. Katia Rodewald: Methodology; investigation; visualization. Bernhard Rieger: Resources. Farah Qoura: Supervision; project administration. Martha Kandawa-Schulz: Writing—review and editing. Norbert Mehlmer: Investigation; supervision. Thomas B. Brück: Conceptualization; methodology; resources; writing—review and editing; supervision; project administration; funding acquisition.
ACKNOWLEDGMENTS
Farah Qoura and Thomas B. Brück would like to acknowledge the financial support of the German Federal Ministry for Economic Affairs and Energy (BMWi) with the grant number 031A305A. Michael Paper was financially supported by the Bavarian State Ministry of the Environment and Consumer Protection within the framework of the ForCycle II Project Group. Nathanael D. Arnold was funded by the German Ministry for Education and Research with grant number 031B0838B in the framework of the BMBF bioeconomy international Canadian/German cooperation project ChitoMat. We would like to express our gratitude to Petrina Kapewangolo and Dirk Woortman for their contribution during the specimen sampling and strain isolation process. The Galaxy server used in this study for some calculations is in part funded by Collaborative Research Centre 992 Medical Epigenetics (DFG grant SFB 992/1 2012) and German Federal Ministry of Education and Research (BMBF grants 031 A538A/A538C RBC, 031L0101B/031L0101C de.NBI-epi, 031L0106 de.STAIR (de.NBI)). Open Access funding enabled and organized by Projekt DEAL.
CONFLICT OF INTEREST STATEMENT
None declared.
ETHICS STATEMENT
None required.
APPENDIX
| Thermosynechococcaceae cyanobacterium sp. Okahandja full 16S ribosomal RNA gene sequence (extracted from genome) ATGGAGAGTTTGATCCTGGCTCAGGATGAACGCTGGCGGTCTGCTTAACACATGCAAGTCGAACGGGCTCTTCGGAGCTAGTGGCGGACGGGTGAGTAACACGTGAGAATCTACCCTTAGGAGGGGGATAACAGTTGGAAACGACTGCTAATACCCCATATGCCGCAAGGTGAAATCTTTTTGGCCTGAGGATGAGCTCGCGGTGGATTAGCTAGTTGGTGGGGTAATGGCCTACCAAGGCAACGATCCATAGCTGGTCTGAGAGGATGATCAGCCACACTGGGACTGAGACACGGCCCAGACTCCTACGGGAGGCAGCAGTGGGGAATTTTCCGCAATGGGCGAAAGCCTGACGGAGCAAGACCGCGTGAGGGACGAAGGCCCTTGGGTTGTAAACCTCTTTTCTCAGGGAAGAACACAATGACGGTACCTGAGGAATAAGCCTCGGCTAACTCCGTGCCAGCAGCCGCGGTAAGACGGAGGAGGCAAGCGTTATCCGGAATTATTGGGCGTAAAGCGTCCGCAGGTGGCTTTCCAAGTCTGCTGTTAAAGCCCGAGGCTCAACCTCGGATCGGCGGTGGAAACTGGATCGCTAGAGTGCGTCAGGGGTAGGGGGAATTCCCGGTGTAGCGGTGAAATGCGTAGATATCGGGAAGAACACCAGCGACGAAAGTGCCCTACTGGGACGTTACTGACACTCATGGACGAAAGCTAGGGGAGCGAAAGGGATTAGATACCCCTGTAGTCCTAGCCGTAAACGATGGACACTAGGTGTTGCTGGTATCCACTCCAGCAGTGCCGTAGCCAACGCGTTAAGTGTCCCGCCTGGGGAGTACGCACGCAAGTGTGAAACTCAAAGGAATTGACGGGGGCCCGCACAAGCGGTGGAGTATGTGGTTTAATTCGATGCAACGCGAAGAACCTTACCAGGGCTTGACATGTCGCGAACCCCGCGGAAACGTGGGGGTGCCTTCGGGAGCGCGAACACAGGTGGTGCATGGCTGTCGTCAGCTCGTGTCGTGAGATGTTGGGTTAAGTCCCGCAACGAGCGCAACCCTCGTCCTTAGTTGCCAGCATTAAGTTGGGCACTCTAGGGAGACTGCCGGTGACAAACCGGAGGAAGGTGGGGATGACGTCAAGTCATCATGCCCCTTACGCCCTGGGCTACACACGTACTACAATGCTGTGGACAGAGAGTTGCCAACCCGCGAGGGCGAGCTAATCTCTTAAACCACGGCTCAGTTCAGATTGCAGGCTGCAACTCGCCTGCATGAAGGCGGAATCGCTAGTAATCGCAGGTCAGCATACTGCGGTGAATACGTTCCCGGGCCTTGTACACACCGCCCGTCACACCATGGGAGTTGGCCACGCCCGAAGTCGTTACTCCAACCCGTAAGGGAGGGGGACGCCTAAGGCAGGGCTGATGACTGGGGTGAAGTCGTAACAAGGTAGCCGTACCGGAAGGTGTGGCTGGATCACCTCCTTT |
| Function | Accession | 50°C log 2FC | 55°C log2 FC | Description |
|---|---|---|---|---|
| CRISPR | 1513 | −2.33 | −4.17 | CRISPR-associated endonuclease Cas1 |
| CRISPR | 1993 | 1.99 | −2.78 | Type I-E CRISPR-associated protein Cas7/Cse4/CasC |
| CRISPR | 1516 | 1.12 | 2.46 | Type III-B CRISPR-associated protein Cas10/Cmr2 |
| Ion Homeostasis/Redox proteins | 1603 | −2.63 | −7.69 | Peroxiredoxin |
| Ion Homeostasis/Redox proteins | 2641 | −4.55 | −6.67 | 2Fe-2S iron-sulfur cluster-binding protein |
| Ion Homeostasis/Redox proteins | 363 | 1.15 | −6.25 | NADH-quinone oxidoreductase subunit J |
| Ion Homeostasis/Redox proteins | 2402 | −1.35 | −4.55 | Peptide-methionine (S)-S-oxide reductase MsrA |
| Ion Homeostasis/Redox proteins | 553 | −2.33 | −4.35 | AarF/ABC1/UbiB kinase family protein |
| Ion Homeostasis/Redox proteins | 1190 | −2.56 | −3.03 | Ferredoxin |
| Ion Homeostasis/Redox proteins | 2070 | 2.37 | −1.43 | Ferredoxin thioredoxin reductase variable alpha chain |
| Ion Homeostasis/Redox proteins | 1860 | 1.09 | 2.27 | SUMF1/EgtB/PvdO family nonheme iron enzyme |
| Ion Homeostasis/Redox proteins | 93 | 1.09 | 2.34 | 2Fe-2S iron-sulfur cluster-binding protein |
| Ion Homeostasis/Redox proteins | 1055 | −1.41 | 2.38 | Ferrous iron transport protein A |
| Metabolism | 2698 | −8.33 | −33.33 | Squalene--hopene cyclase |
| Metabolism | 1725 | −5.88 | −14.29 | TldD/PmbA family protein |
| Metabolism | 2289 | −1.92 | −11.11 | Phosphoribosylglycinamide formyltransferase |
| Metabolism | 190 | −6.25 | −9.09 | M23 family metallopeptidase |
| Metabolism | 2040 | −5.88 | −7.69 | Glucose-6-phosphate isomerase |
| Metabolism | 955 | −3.23 | −7.14 | Citramalate synthase |
| Metabolism | 2427 | −1.18 | −5.88 | Glycosyltransferase family 1 protein |
| Metabolism | 1135 | −5.00 | −5.88 | Haloacid dehalogenase superfamily, subfamily IA |
| Metabolism | 2708 | −2.38 | −5.00 | Glycosyltransferase |
| Metabolism | 2533 | −4.55 | −4.76 | Deoxyhypusine synthase |
| Metabolism | 2677 | −1.89 | −4.35 | PFAM Glyoxalase bleomycin resistance protein dioxygenase |
| Metabolism | 1170 | −3.33 | −4.17 | Chorismate mutase |
| Metabolism | 2479 | −1.67 | −4.00 | NUDIX hydrolase |
| Metabolism | 312 | −1.05 | −3.85 | Bifunctional pantoate--beta-alanine ligase/(d)CMP kinase |
| Metabolism | 917 | 1.35 | 2.17 | fructosamine kinase family protein |
| Metabolism | 104 | −1.49 | 3.54 | GNAT family N-acetyltransferase |
| Photosynthesis | 1482 | −4.17 | −9.09 | HEAT repeat domain-containing protein |
| Photosynthesis | 623 | −8.33 | −7.69 | Protoporphyrinogen oxidase |
| Photosynthesis | 2629 | −4.76 | −6.67 | Apocytochrome f |
| Photosynthesis | 694 | −2.17 | −6.25 | Photosystem I reaction center subunit II |
| Photosynthesis | 1748 | −2.70 | −5.56 | Chlorophyll a-b binding domain-containing protein |
| Photosynthesis | 1572 | −2.04 | −3.57 | Photosystem II manganese-stabilizing polypeptide |
| Photosynthesis | 1413 | −1.05 | −2.63 | Photosynthetic/respiratory NAD(P)H-quinone oxidoreductase subunit C |
| Photosynthesis | 697 | −1.14 | −2.22 | Chlororespiratory reduction protein 7 |
| Photosynthesis | 1483 | 1.16 | 3.15 | Phycobilisome rod-core linker polypeptide |
| Regulators | 652 | −5.26 | −25.00 | YciI family protein |
| Regulators | 1449 | −4.76 | −16.67 | YbaB/EbfC family nucleoid-associated protein |
| Regulators | 302 | −2.86 | −8.33 | ATP-binding protein |
| Regulators | 1885 | −1.35 | −6.67 | GGDEF domain |
| Regulators | 1377 | −2.38 | −5.26 | Metalloregulator ArsR/SmtB family transcription factor |
| Regulators | 2557 | −3.03 | −4.55 | Heat-inducible transcriptional repressor HrcA |
| Regulators | 1647 | −2.17 | −3.70 | LysR family transcriptional regulator |
| Regulators | 1886 | −2.22 | −2.33 | Circadian clock protein KaiA |
| Regulators | 2313 | −3.85 | −1.69 | Translation elongation factor 4 |
| Regulators | 1693 | 1.28 | 3.22 | Response regulator |
| Regulators | 960 | 1.79 | 11.01 | Signal Transduction Histidine Kinase |
| Transport | 1624 | −4.00 | −6.67 | Sulfate ABC transporter permease subunit CysT |
| Transport | 2077 | −2.94 | −4.00 | Energy-coupling factor ABC transporter permease |
| Transport | 710 | −2.08 | −3.03 | Protein translocase subunit SecF |
| Transport | 506 | −1.14 | −2.86 | LPS export ABC transporter ATP-binding protein |
| Transport | 908 | −1.14 | −2.38 | DUF4040 domain-containing protein |
| Transport | 1681 | 1.7 | 2.79 | SLC13 family permease |
| Transport | 1171 | 1 | 3.23 | Signal peptide peptidase SppA |
| Transport | 100 | 2.54 | 4.58 | Carbohydrate ABC transporter permease |
| Genetic replication | 2324 | −1.82 | −6.67 | DNA adenine methylase |
| Genetic replication | 906 | −4.00 | −5.88 | Segregation/condensation protein A |
| Genetic replication | 158 | −2.44 | −5.00 | 30S ribosomal protein S12 |
| Genetic replication | 418 | 1.11 | −3.45 | Pentapeptide repeat-containing protein |
| Genetic replication | 594 | −2.44 | −3.45 | Polyribonucleotide nucleotidyltransferase |
| Genetic replication | 2513 | −2.38 | −3.13 | DNA-3-methyladenine glycosylase |
| Genetic replication | 2297 | −3.33 | −2.94 | Ribosome-associated translation inhibitor RaiA |
| Genetic replication | 2574 | −2.70 | −2.86 | Ribonuclease H-like domain-containing protein |
| Genetic replication | 2711 | 1.53 | −2.78 | Hypothetical protein pgaptmp_002711 |
| Genetic replication | 1611 | 1.16 | 3.02 | DNA polymerase III subunit delta' |
| Genetic replication | 421 | 1.13 | 4.3 | Phenylalanine--tRNA ligase subunit beta |
| Genetic replication | 2046 | 2.72 | 5.79 | Crossover junction endodeoxyribonuclease RuvC |
| Genetic replication | 2673 | 1.23 | 7.13 | 50S ribosomal protein L33 |
| Genetic replication | 2469 | 3.03 | 13.13 | Hypothetical protein pgaptmp_002469 |
- Upregulated at:
 55°C only
55°C only  50°C and 55°C,
50°C and 55°C,  50°C only
50°C only - Downregulated at:
 55°C only,
55°C only,  50°C and 55°C increasingly,
50°C and 55°C increasingly,  50°C more, 55° C less again.
50°C more, 55° C less again.

Open Research
DATA AVAILABILITY STATEMENT
All relevant data are provided in full in the results and appendix section of this paper. Additional analyses and raw data sets can be accessed in the Zenodo repository at https://zenodo.org/doi/10.5281/zenodo.10007199. The Thermosynechococcaceae cyanobacterium sp. Okahandja genome can be found at NCBI under accession CP136945. The 16S rRNA gene sequence can be accessed at GenBank under OR689577 or in Appendix Table A1.