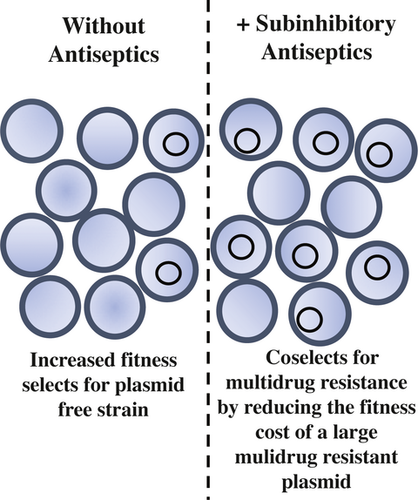
Fitness costs associated with carriage of a large staphylococcal plasmid are reduced by subinhibitory concentrations of antiseptics
Graphical Abstract
Pathogenic bacteria often carry large plasmids that contain multiple resistance genes; individually, these elements coselect for additional resistance genes that are genetically linked by the plasmid. In this short communication, we demonstrate increased triclosan resistance can be conjugatively transferred in Staphylococcus aureus. Furthermore, in a competition assay, we show subinhibitory concentrations of antiseptics abrogate the fitness cost of a large multidrug resistance plasmid in S. aureus, thus coselecting for the numerous antimicrobial resistance genes present on the plasmid.
Abstract
Staphylococcus aureus carries a collection of mobile genetic elements that often harbor virulence and antimicrobial resistance genes. Since the introduction of antibiotics, plasmids have become a major genetic element responsible for the distribution of antimicrobial resistance. Under antimicrobial selection, resistance plasmids are maintained within bacterial populations as a means to ensure survival. However, in the absence of selection, large plasmids can be lost due to the fitness costs associated with harboring these genetic elements. pC02 is a previously identified multidrug resistance, conjugative plasmid that is found in S. aureus. In addition to antibiotic resistance, pC02 also carries genes known to be associated with antiseptic resistance. Among these, we previously characterized the contribution of qacA to pC02 mediated reduced chlorhexidine susceptibility. Herein, we demonstrate that pC02 also mediates triclosan resistance, likely due to the presence of fabI, a known triclosan resistance gene. Moreover, we demonstrate that conjugative transfer of pC02 increases triclosan resistance in recipient cells. Competition assays demonstrated a fitness cost associated with carriage of the large pC02 plasmid. However, subinhibitory concentrations of either chlorhexidine or triclosan abrogated this fitness cost. Given the widespread use of these antiseptics, both of which accumulate in wastewater and other environmental reservoirs, indiscriminate use of antiseptics likely imposes a constant selective pressure that promotes maintenance of antimicrobial resistance factors within S. aureus.
1 INTRODUCTION
Infections caused by multidrug-resistant S. aureus continue to result in therapeutic failure and extended hospital stays. The evolution of multidrug resistance in bacteria is associated with nonessential antibiotic applications and improper administration of these agents (Andersson and Hughes, 2010; Foster, 2017; Munita & Arias, 2016; Ventola, 2015). Today, it is not uncommon for pathogens to be resistant to multiple antibiotics. Antibiotic resistance is commonly attributed to large plasmids, which have accumulated resistance genes over time. In addition to antibiotic resistance genes, plasmids can also harbor biocide and heavy metal resistance genes (McCarthy & Lindsay, 2012). When found on the same genetic element, this collection of resistance genes can provide protection against a wide variety of compounds, any one of which can provide a selective pressure to maintain the entire genetic element; this phenomenon is known as coselection (Gullberg, Albrecht, Karlsson, Sandegren, & Andersson, 2014; Pal, Bengtsson-Palme, Kristiansson, & Larsson, 2015). Given the increased awareness of the need to reduce unnecessary antibiotic prescriptions, some coselective pressures are being minimized. However, increased utilization of antiseptics, such as quaternary ammonium compounds and chlorhexidine, in common use household products have increased their abundance in environmental reservoirs. To this end, little is known about the coselective pressures imposed by subinhibitory concentrations of antiseptic compounds on multidrug resistance plasmids in S. aureus.
In our previous work, we identified and characterized a large multidrug resistance conjugative plasmid in S. aureus called pC02 (Johnson et al., 2015; LaBreck, Li, Gibbons, & Merrell, 2019; LaBreck et al., 2018). In the prior characterization study, pC02 was shown to be a fusion between at least two theta replicating plasmids: one plasmid containing the conjugative gene cluster and the other harboring antibiotic resistance genes (erythromycin, beta-lactam resistance), a heavy metal resistance gene (cadmium resistance), and biocide resistance genes (LaBreck et al., 2019). For biocide resistance, pC02 harbors qacA and fabI, which offer reduced susceptibility to chlorhexidine and triclosan, respectively. We previously demonstrated that pC02 qacA mediates reduced susceptibility to chlorhexidine (LaBreck et al., 2018). Herein, we demonstrate that pC02 also mediates reduced triclosan susceptibility. To further dissect the implications of carrying these biocide resistance genes, we analyzed the potential of subinhibitory concentrations of antiseptics to coselect for other resistance genes on pC02.
2 MATERIALS AND METHODS
2.1 Bacteria and growth conditions
Staphylococcus aureus 2014.C02 (S. aureus C02), S. aureus C02-M2, S. aureus C02-RN, and S. aureus RN TC (pC02F) were previously described (LaBreck et al., 2019, 2018). Briefly, S. aureus C02 harbors the conjugative plasmid pC02 (GenBank: CP012121.2). S. aureus C02-M2 is an isogenic variant of S. aureus C02 that has been cured of pC02. S. aureus C02-RN is a spontaneously resistant rifampin and novobiocin-resistant strain that was derived in the S. aureus C02-M2 background. S. aureus C02-RN TC (pC02F) is a transconjugant derivative of a mating between S. aureus C02 and C02-RN wherein C02-RN obtained pC02 by conjugation. The strains are archived in our strain collection under the following DSM numbers: S. aureus 2014.C02 (DSM 1418), S. aureus C02-M2 (DSM 1552), S. aureus C02-RN (DSM1553), and S. aureus C02-RN TC (pC02F) (DSM1699). All strains were grown in Mueller Hinton Broth II (Becton Dickinson). For solid media, MHB was supplemented with 1.7% agar.
2.2 Triclosan susceptibility testing
The MIC of triclosan was tested in a macrobroth dilution assay as previously described (Johnson et al., 2015; LaBreck et al., 2018) with minor modifications. Briefly, approximately 5.0 × 104 colony forming units (CFU) were inoculated into 2 ml of MHB containing increasing concentrations (0.2 μg/ml to 1.0 μg/ml in increments of 0.2 μg/ml, or 0.002 μg/ml to 0.01 μg/ml in increments of 0.002 μg/ml) of purified (≥97.0%) Irgasan (Sigma). The cultures were grown at 37°C degrees overnight, shaking at 220 rpm. The MIC was determined by visual analysis of the culture's turbidity. Similarly, the MIC of chlorhexidine for C02-RN was tested by microbroth dilution as previously described (Johnson et al., 2015; LaBreck et al., 2018).
2.3 pC02 fitness cost
Strains of C02 and C02-M2 (C02 cured of pC02) were grown overnight in 2 ml of Muller Hinton Broth (MHB). The following day, approximately 2.5 × 104 CFU of each strain was combined in 2 ml of MHB. About 10 µl was taken from the mixed culture for input CFU determination, and the culture was grown shaking at 37°C for 24 hr. CFU for C02 were determined by plating on MHA supplemented with cadmium chloride; CFU for C02-M2 were determined by plating on MHA followed by subtraction of the total CFU determined for C02. The following day, CFU for C02 and C02-M2 were determined from the 24-hr mixed culture, and the culture was passaged as approximately 1 × 105 CFU into 2 ml of fresh MHB. The culture was repeatedly passaged as described above, and CFU were determined at 48, 72, and 96 hr as a means to determine the competitive fitness, as represented by a competition index, of C02 and C02-M2 over time. A similar protocol was followed for experiments that were conducted with subinhibitory concentrations of antiseptics with minor modifications. Briefly, strains were grown overnight without selection in MHB, the following day equal CFU of the strains were added to 2 ml of MHB supplemented with either 0.1 μg/ml, 0.5 μg/ml, or 0.01 μg/ml chlorhexidine or 0.003 μg/ml, 0.0015 μg/ml, or 0.00075 μg/ml triclosan. Antiseptic selection was maintained throughout the multiple passages. The spread in the gathered data was likely attributed to the plating method used to determine total CFU of the bacteria; 10 μl streak dilutions were used. The subinhibitory antiseptic concentrations were empirically selected based on strain MIC values, which are shown in Table 1 (Buffet-Bataillon, Tattevin, Bonnaure-Mallet, & Jolivet-Gougeon, 2012).
| Strain | Triclosan | Chlorhexidinea | ||
|---|---|---|---|---|
| Medianb | Max | Median | Max | |
| C02 | 1 | 1 | 0.9 | 1 |
| C02-M2 | 0.006 | 0.006 | 0.3 | 0.4 |
| C02-RN | 0.008 | 0.01 | 0.2 | 0.2 |
| C02-RN TC (pC02F) | 1 | 1 | 0.5 | 0.5 |
- a Data for chlorhexidine MIC for C02, C02-M2, and C02-RN TC (pC02F) are taken from LaBreck et al. (2018).
- b Values are concentrations in µg/ml. Data were obtained from at least 3 biologically independent experiments.
The competitive index was calculated using the following formula: Output (CFU C02-M2/CFU C02)/Input (CFU C02-M2/CFU C02).
3 RESULTS AND DISCUSSION
On pC02, fabI is found on the TnSha1 element (Furi et al., 2016) (Figure 1). To determine the extent of triclosan resistance mediated by this element, we measured the triclosan MIC of strain C02 and a plasmid cured variant of C02, C02-M2, in a macrobroth dilution assay. The MIC of C02 was >100-fold higher than C02-M2, suggesting that the fabI resistance gene found on pC02 offered substantial protection from triclosan (Table 1). As pC02 is a conjugative plasmid, we next sought to determine whether this resistance was transferrable. To this end, we examined the MICs of triclosan for a recipient strain as well as a pC02 transconjugant in the same strain background. Similarly, we identified a >100-fold increase in triclosan resistance (Table 1). Together, these data indicated that pC02 provided significant protection against triclosan and that this resistance was transferrable by conjugation.
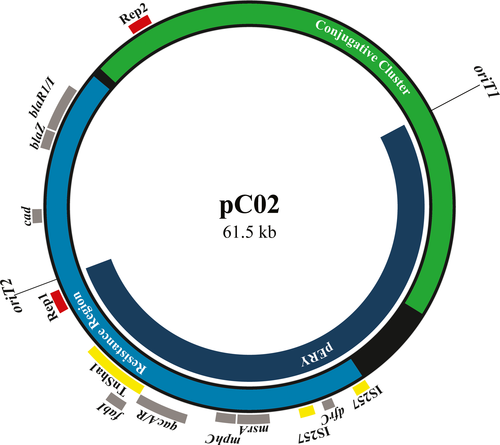
The presence of two functional antiseptic resistance genes (qacA and fabI) on pC02 suggests that S. aureus is under increasing selective pressure from exposure to different antiseptic agents. Interestingly, these genes are adjacent to each other, which may increase the frequency of cotransfer via conjugation. To this end, in addition to being found on the fully conjugated pC02, both fabI and qacA are found on pERY (a partially transferred pC02 exconjugant plasmid) (Figure 1), which we previously demonstrated displayed an increased conjugative transfer frequency as compared to pC02 (LaBreck et al., 2019). However, the partially transferred pERY is unstable without selection, likely nonconjugative, and similar plasmids have not been deposited into the NCBI database (LaBreck et al., 2019). These data suggest the pERY plasmid variant is uncommon in nature if it exists at all. Furthermore, it is unlikely that qacA and its regulator qacR move with fabI on the TnSha1 element; no IS1272-associated inverted repeats are found near qacA or qacR (data not shown). Regardless, the presence of qacA, fabI, and additional resistance elements provides an example of resistance gene clustering on a plasmid.
The resistance cluster on pC02 is located within a region that encompasses ~45% of the total plasmid sequence (Figure 1). This region and the conjugative cluster comprise the 61,539 bp sequence of pC02, which is the 5th largest S. aureus plasmid in the NCBI database. Given the large size of pC02, we hypothesized that there could be a fitness cost associated with carriage of pC02. To assess the fitness cost of pC02, we determined the competitive index of C02 versus C02-M2 (pC02-cured C02) in a daily passaged mixed culture assay. In the absence of antimicrobial agents, there was a >10-fold fitness advantage for C02-M2 after 48 hr, and this difference was further exaggerated at 72 and 96 hr (Figure 2a). These data indicate that pC02 impacts host fitness in the examined growth conditions.
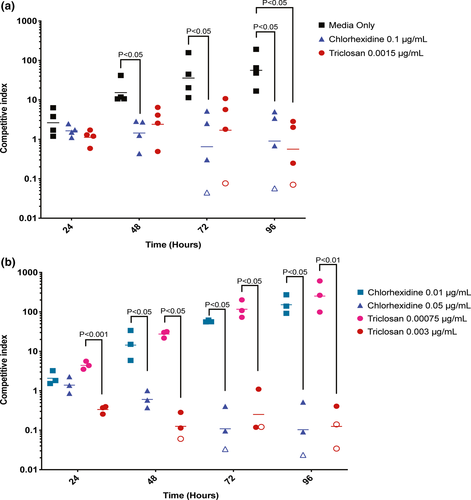
Antiseptic compounds are used daily in hospitals and communities. After their application, the majority of antiseptic compounds are disposed of in wastewater and then accumulate at subinhibitory concentrations in the environment (Mulder et al., 2018; Zhang et al., 2015). To this end, subinhibitory concentrations of chlorhexidine in sludge and influent wastewater have been measured at 7.6 μg/ml and 0.001185 μg/ml, respectively (Östman, Lindberg, Fick, Björn, & Tysklind, 2017). Similarly, triclosan has been shown to be present in influent wastewater at concentrations of 0.0038–0.0166 μg/ml (McAvoy, Schatowitz, Jacob, Hauk, & Eckhoff, 2002). To determine whether subinhibitory concentrations of antiseptics could reduce the relative fitness cost of pC02, we repeated the mixed culture experiment in subinhibitory concentrations of two antiseptics. In the presence of subinhibitory chlorhexidine (0.1 μg/ml) or triclosan (0.0015 μg/ml), the fitness cost of harboring pC02 was partially or fully negated throughout the entire 96 hr (Figure 2a). To determine whether the phenotype was similar in different concentrations of subinhibitory chlorhexidine and triclosan, we tested two additional concentrations for each antiseptic (Figure 2b). Again, the fitness cost associated with pC02 carriage was ameliorated in 0.05 μg/ml chlorhexidine or 0.003 μg/ml triclosan. However, this amelioration was not seen in chlorhexidine and triclosan at 0.01 μg/ml and 0.00075 μg/ml, respectively (Figure 2b); thus, a minimum concentration threshold of each antiseptic was required to eliminate the fitness cost associated with carriage of pC02. Together, these data indicate that biologically relevant subinhibitory concentrations of antiseptic compounds can select for strains that carry antiseptic resistance plasmids.
Antiseptics have become an integral component in many healthcare and common use consumer products. When applied at recommended concentrations, they are effective at inhibiting microbial contamination. However, given their increasing applications, antiseptic compounds have inevitably accumulated in the environment at subinhibitory concentrations (Mulder et al., 2018; Zhang et al., 2015). Toxicities from QACs and triclosan have been demonstrated for various aquatic organisms (Dann & Hontela, 2011; Di Nica, Gallet, Villa, & Mezzanotte, 2017; Zhang et al., 2015). However, little is known about the effects of subinhibitory concentrations of antiseptics in wastewater. Other groups have demonstrated increased plasmid stability in subinhibitory concentrations of metals and antiseptics, as well as cross-resistance development in subinhibitory antibiotic concentrations (Braoudaki & Hilton, 2004; Buffet-Bataillon et al., 2012; Gullberg et al., 2014; Kampf, 2018; Wand, Bock, Bonney, & Sutton, 2017). Outside of the aquatic environment, subinhibitory concentrations of antiseptics can also be found on surfaces, which provide similar selective pressures (Bashir et al., 2019; Ribič, Klančnik, & Jeršek, 2017). Herein, we showed that subinhibitory concentrations of antiseptics negated the fitness cost associated with carrying a large plasmid in S. aureus (Figure 2), and in doing so coselected for the presence of multiple antimicrobial resistance genes on pC02. En masse, the data support the need for greater antiseptic stewardship.
ACKNOWLEDGMENTS
These studies were supported by a U.S. Department of Defense Program project grant (#HT9404-12-1-0019) and a Military Infectious Diseases Research Program award (#HU0001-15-2-0031).
CONFLICT OF INTEREST
The contents of this publication are the sole responsibility of the author(s) and do not necessarily reflect the views, opinions, or policies of Uniformed Services University, the Henry M. Jackson Foundation for the Advancement of Military Medicine, nor the U.S. Government. DSM is an employee of the U.S. Government. Title 17 U.S.C. 105 provides that “Copyright protection under this title is not available for any work of the United States Government.”
AUTHOR CONTRIBUTION
Patrick T LaBreck: Investigation; Methodology; Writing-original draft; Writing-review & editing. D. Scott Merrell: Investigation; Methodology; Resources; Writing-original draft; Writing-review & editing.
ETHICS STATEMENT
None required.
Open Research
DATA AVAILABILITY STATEMENT
All data are provided in the manuscript. Original raw data are available by request. GenBank accession numbers for S. aureus 2014.C02 and pC02 are CP012120.2 (https://www.ncbi.nlm.nih.gov/nuccore/CP012120.2) and CP012121.2 (https://www.ncbi.nlm.nih.gov/nuccore/CP012121.2), respectively.