Colorimetric detection of SARS-CoV-2 variants with paper-based analytical devices
Graphical Abstract
Given that the severe acute respiratory syndrome coronavirus 2 (SRAS-CoV-2) virus keeps mutating during the COVID-19 pandemic, there is an urgent need to develop a quick and convenient test method to detect real-life samples for mass on-site screening. Zhang et al. proposed a novel strategy for rapid detection and discrimination of SARS-CoV-2 and its variants at rather low cost utilizing a subtly designed DNA probe. By combining with a paper-based colorimetric amplification platform, the results could be analyzed by the naked eyes or a widely used smartphone.
Recently, Zhang et al.1 reported a colorimetric and paper-based assay for rapid detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its variants leveraging a DNA probe-mediated specific nucleic acid strand-displacement reaction and urease-based colorimetric readout. This assay offered a low-cost, easy-to-use, efficient, and scalable diagnosis strategy for the detection of SARS-CoV-2 and its variants.
Since December 2019, the SARS-CoV-2 virus, responsible for the rapid and uncontrollable worldwide outbreak of coronavirus disease, has spread unscrupulously virtually in every corner of the world, posing a severe threat to human health and the global economy.1 To reduce the transmission of SARS-CoV-2, accurate viral detection is an indispensable step before the isolation and treatment of positive individuals. At present, the main principle of detection is to analyze the nucleic acid of the virus,2 by which quantitative real-time polymerase chain reaction with reverse transcription (RT-qPCR) is leveraged as the gold-standard method for SARS-CoV-2 detection due to its high accuracy and fidelity.2 But it is time-consuming, expensive, and relies on laboratory instruments and specialists, limiting its wide use in low-resource areas. On the other hand, isothermal nucleic acid amplification is an efficient alternative.1 For instance, reverse transcription loop-mediated isothermal amplification, transcription-mediated amplification, and CRISPR-based detection techniques are developed for rapid, convenient, and efficient detection of virus RNA, simplifying the instruments and procedures (Supporting Information: Table S1). Nevertheless, these emerging techniques are not mature enough to face the diagnostic challenges associated with virus evolution. The newly emerging SARS-CoV-2 variants with the features of higher transmissibility and virulence bring a greater challenge to diagnosis.3 Therefore, developing a rapid and easy-to-use method is a matter of great urgency for the simultaneous detection of SARS-CoV-2 and its variants. Now, writing in Nature Biomedical Engineering, Zhang et al. reported a low-cost (about US $0.30 per test), paper-based assay for rapid (about 30 min per test) detection of SARS-CoV-2 and its variants just via naked eyes or a smartphone, obviating the utilization of specialized equipments.1
This assay, which they termed MARVE (abbreviation for multiplexed, nucleic-acid-amplification-free, single-nucleotide-resolved viral evolution), utilized a highly sensitive and selective DNA probe for specific recognition of viral mutated RNA (SARS-CoV-2 variants Alpha, Beta, Gamma, and Delta) and urease enzymes to offer a visible colorimetric signal (Figure 1A). The DNA probe, that is, the toehold exchange DNA probe (TEprobe), consisted of two partially complementary DNA strands labeled with Rec and Block, respectively. There was a pair of terminal toehold regions in TEprobe, referred to as the forward and reverse toehold, which could precisely control the strand displacement reaction induced by input RNAs. Also, a cytosine–cytosine (C:C) mismatch site was incorporated within the reverse toehold region to mediate the formation of a C-Ag(I)-C metal-mediated base pair via base–metal cation interactions. With such an elegant design, the target viral RNA could competitively bind to the Rec strand and replace the Block strand via the forward toehold-mediated strand displacement reaction, leading to the release of Ag(I) ions which subsequently inhibited the activity of urease. Thereafter, inactivated urease hindered the hydrolysis of urea to NH4+, which could not change the original pH, offering a yellow readout in the presence of phenol red (which is a widely available pH indicator). Conversely, nontargeting RNAs could not effectively replace the Block strand and then hindered the release of Ag(I), offering a red readout.
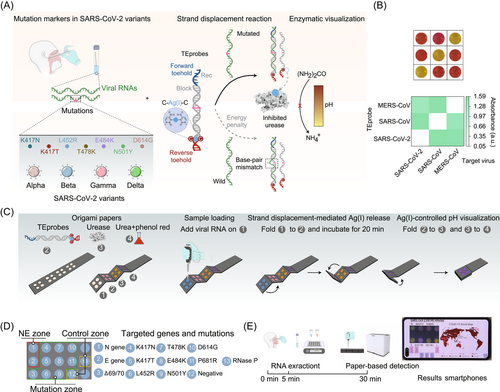
Based on the principles described above, Zhang et al. first tested the capacity of MARVE in detecting and discriminating several respiratory viruses. By optimizing different TEprobes, MARVE gave a sensitive platform that could specifically detect and discriminate three coronaviruses, including SARS-CoV, Middle East respiratory syndrome coronavirus, and SARS-CoV-2 (Figure 1B). And seven influenza virus subtypes, which can cause similar symptoms to SARS-CoV-2, could also give clear color distinctions based on MARVE. Then, to deal with an increasing need for mass on-site screening, a creative strategy was developed by coupling origami papers with the reactions of MARVE to offer a simple and rapid diagnostic platform. The origami papers, consisting of four paper layers, were designed by placing the samples on the bottom layer, which was subsequently folded up and the layer on the top gave a colorimetric readout (Figure 1C). In addition, on account of the scalability of origami papers and the programmability of probes, multiple targets would be distinguished.
Next, the MARVE assay was utilized to detect four SARS-CoV-2 variants including Alpha, Beta, Gamma, and Delta. The TEprobes were optimized to target specific mutated RNAs, with which SARS-CoV-2 and its variants could be readily discriminated. Because of the expandability of origami papers, nine mutations of SARS-CoV-2 variants could be integrated into one test paper, satisfying the need for diagnosing multiple variants (Figure 1D). In addition, MARVE could test samples from an optimized thermal lysis process to facilitate rapid RNA extraction, avoiding cross contaminations and amplification/enrichment processes (Figure 1E). To accommodate the rapid evolution of the SARS-CoV-2 virus and detection requirements for emerging variants, a general method was devised to customize MARVE for detecting emerging variants. Especially, with widespread availability and mobility, smartphones can be a favorable tool in the field of imaging, analysis, and transmission of results. Therefore, the analyzed results could be interpreted by smartphone-based customized software, eliminating the need for bulky optical inspection instruments.
Finally, they also demonstrated the feasibility of MARVE for analyzing clinical samples. 50 clinical throat swab samples (35 positive samples and 15 negative samples) were tested in parallel by RT-PCR and MARVE assays. Impressively, the results obtained by both protocols were completely consistent. Of note, they also sequenced the RNAs of positive samples with a D614G mutation and utilized MARVE to validate the existence of the D614G mutation.
Overall, Zhang et al. substantiated that MARVE could be a feasible tool for rapid discrimination of SARS-CoV-2 variants at rather low cost. They made use of special TEprobes to discriminate SARS-CoV-2 variants and urease enzymes to offer a visual readout. Paper-based colorimetry is considered a promising strategy for rapid and on-site detection of multiple analytes at the same time. Confining the probes and reactions on the origami paper allowed the simultaneous analysis of multiple variants, where the results could be read by eyes or a smartphone while offering quantitative colorimetric analysis. Considering the large-scale transmission of SARS-CoV-2, paper-based MARVE would provide a possible alternative platform for rapid, large-scale, and out-of-lab screening of COVID-19-positive individuals at an early stage, thereby timely preventing further spread of the virus.
MARVE is an efficient tool to meet the challenge of the COVID-19 pandemic. However, there is still room in enhancing the sensitivity of MARVE to match the current state-of-the-art RT-qPCR technology. Otherwise, diagnosis lapses would lead to the spread of infectious diseases by false-negative individuals. Modification of electrodes on paper might be a promising strategy to improve the sensitivity of signal acquisition by detecting current changes.4 Chemical modification of paper surface is a realistic strategy by improving the colorimetric performances of paper-based devices, such as the generated color intensity and uniformity, to improve test sensitivity.5 At present, this assay still cannot avoid the employment of lysis devices for RNA extraction, however, a compact integrated device is an irresistible trend. Hence, the simplification of the test procedure and device to an integrated system needs to be further explored.
AUTHOR CONTRIBUTIONS
Qi Wang and Zhaowei Chen drafted the manuscript. Huanghao Yang revised the manuscript. All authors have read and approved the final manuscript.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2020YFA0210800, Huanghao Yang), the National Natural Science Foundation of China (22027805, Huanghao Yang; 22277011, Zhaowei Chen; 22107019, Zhaowei Chen), and the Major Project of Science and Technology of Fujian Province (2020HZ06006, Huanghao Yang).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ETHICS STATEMENT
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
Not applicable.