Automated quantitative analysis of multiple cardiomyocytes at the single-cell level with three-dimensional holographic imaging informatics
Abstract
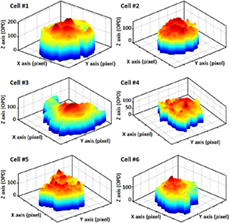
Cardiomyocytes derived from human pluripotent stem cells are a promising tool for disease modeling, drug compound testing, and cardiac toxicity screening. Bio-image segmentation is a prerequisite step in cardiomyocyte image analysis by digital holography (DH) in microscopic configuration and has provided satisfactory results. In this study, we quantified multiple cardiac cells from segmented 3-dimensional DH images at the single-cell level and measured multiple parameters describing the beating profile of each individual cell. The beating profile is extracted by monitoring dry-mass distribution during the mechanical contraction-relaxation activity caused by cardiac action potential. We present a robust two-step segmentation method for cardiomyocyte low-contrast image segmentation based on region and edge information. The segmented single-cell contains mostly the nucleus of the cell since it is the best part of the cardiac cell, which can be perfectly segmented. Clustering accuracy was assessed by a silhouette index evaluation for k-means clustering and the Dice similarity coefficient (DSC) of the final segmented image. 3D representation of single of cardiomyocytes. The cell contains mostly the nucleus section and a small area of cytoplasm.