Organoid-driven diagrammatic devolution: Elevating precision in pancreatic cystic lesions diagnosis
Fei Jiang and Dongyan Cao contributed equally to this work.
Dear Editor,
Pancreatic cystic lesions pose a diagnostic challenge. The accuracy of distinguishing low-grade from high-grade dysplasia is suboptimal, with the progression risk varying based on the types of cysts (simple retention cysts, pseudocysts and cystic neoplasms).1 Traditional imaging-based radiological approaches (computed tomography [CT] and magnetic resonance imaging [MRI]), endoscopic ultrasound (EUS)-guided fine needle aspiration, including analysis of cystic fluid components such as amylase, glucose, carcinoembryonic antigen (CEA) levels, liquid-based cytology and more recently molecular markers can enhance the diagnosis of pancreatic cystic lesions.2 Even though, there is a pressing need for a stable and accurate model that allows in-depth analysis of cell components of cystic lesions and reflects their behaviour. Organoids as 3D multicellular structures resemble features of their original tissue individually for self-organization and self-renewal.3, 4 Organoid-based longitudinal testing aids in monitoring translational diagnosis, disease progression, treatment response and adapting therapies. To enhance precision in diagnosing pancreatic cystic lesions, we collected cystic fluids for organoid culture, evaluating their growth phenotypes and molecular markers. The cell context of successfully constructed organoids was further validated by single-cell RNA sequencing (scRNA-Seq) (Figure 1A).
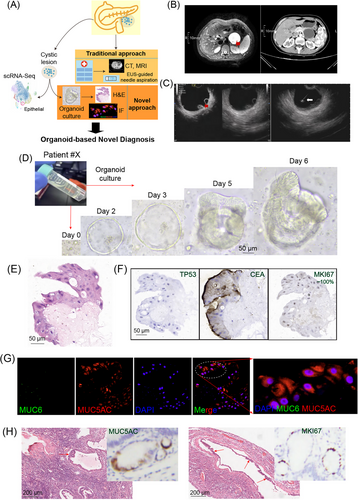
A comprehensive explanation of the methods is in the Supporting Information. A 34-year-old female (patient #X) complained of epigastric pain persisting for 5 years with recurrent pancreatitis. The CT data showed a pancreatic cystic lesion measuring 5 cm in diameter within the pancreatic body, raising suspicion of a pancreatic pseudocyst (Figure 1B). Subsequent enhanced MRI and contrast EUS suggested the lesion as a mucinous cystic neoplasm with enhanced mural nodules (Figure 1C). An EUS-guided fine needle aspiration was conducted and approximately 40 mL of cystic fluid was aspirated. The cystic fluid revealed amylase at a level of 165 328 U/L (>250 U/mL suggests the possibility of pancreatic pseudocyst), CEA level of 261.98 ng/mL (>192 ng/mL indicates the possibility of pancreatic mucinous cystic neoplasm) and glucose level of 9 mg/dL (<50 mg/dL suggests the possibility of pancreatic mucinous cystic neoplasm). Meanwhile, a small volume of cyst fluid (∼3 mL) from the puncture was performed for organoid culture (Figure 1D). Notably, organoids derived from this case exhibited robust growth. Haematoxylin and eosin staining highlighted abnormal structures in cell nuclei (Figure 1E). Immunohistochemical staining for CEA, TP53 and MIK67, as well as immunostaining for MUC5AC, all yielded positive results (Figure 1F,G). These staining data may confirm the diagnosis of this patient with high-grade intraepithelial neoplasia.
The patient eventually underwent surgical resection because of recurrent episodes of acute pancreatitis, and postoperative pathology reported pancreatic mucinous cystadenoma with low-grade intraepithelial neoplasia. This discrepancy raised concerns among the authors and our medical team, prompting us to question whether incomplete sampling during postoperative pathology could have influenced the findings. Experienced pathologists were consulted to re-slice, stain and review additional wax blocks, ultimately discovering evidence of high-grade lesions in certain areas (as indicated by arrows in Figure 1H), along with positive staining of MUC5AC and MKI67 (Figure 1H). These findings corroborated the initial organoid culture results, confirming the presence of high-grade intraepithelial neoplasia.
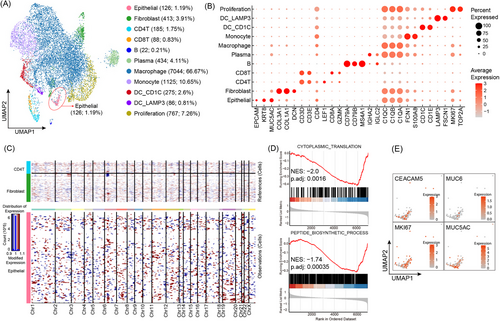
In addition, to validate the accuracy of diagnosis for patient #X at the molecular level, about 3 mL of cystic fluid was subjected to scRNA-Seq. After quality control filtering, 10 565 single cells were retained for subsequent analysis (Figure 2A).5 A total of 11 cell types were identified. They are fibroblast (COL1A1, COL3A1 and DCN), CD4T (CD3D, CD3E and CD4), CD8T (CD3D, CD3E and CD8A), B (CD79A and MS4A1), plasma (IGHA2 and IGLC2), macrophage (C1QC, C1QB and C1QA), monocyte (FCN1 and S100A8), DC_CD1C (CD1C and CD1E), DC_LAMP3 (LAMP3 and FSCN1), proliferation (MKI67 and TOP2A) cells and importantly an epithelial (EPCAM, KRT8 and MUC5AC) group (Figure 2B). The copy number variation (CNV) levels of epithelial cells vary (Figure 2C). Gene set enrichment analysis indicated that the CNV_low group was enriched in cytoplasmic translation and peptide biosynthetic processes (Figure 2D). The RNA expression patterns, as revealed by the scRNA-Seq data, showed concordance with the protein levels (Figure 2E). These findings align with the organoid data, supporting the diagnosis of high-grade over low-grade intraepithelial neoplasia.
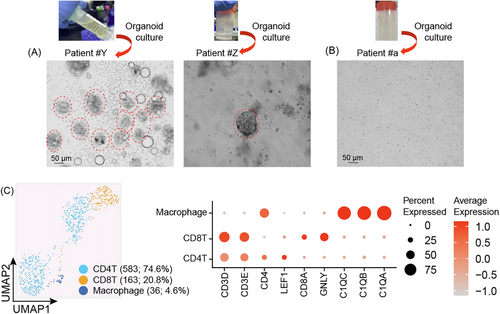
We expanded our study by including three additional patients with pancreatic cystic liquid to assess the viability of organoid technology as a novel and functionally advanced diagnostic approach for this disease. Pancreatic cystic liquid from patients #Y and #Z diagnosed with high-grade intraepithelial neoplasia underwent organoid culture, and organoids were promptly observed (Figure 3A). In contrast, cystic liquid from patient #a with low-grade culture did not yield organoids (Figure 3B). This finding is in concordance with scRNA-Seq data, indicating the absence of epithelial cells (Figure 3C). The dominant populations in the cystic fluid were identified as CD4T, CD8T and macrophages. These organoids provide a comprehensive view of pancreatic cystic lesions, crucial for diagnosing their grade.
Here, we collect pancreatic cystic fluid and culture it to generate organoids. This methodology helps to functionally validate the distinction between cystic lesions with low-grade and patients with high-grade dysplasia, thereby aiding subsequent pathological diagnoses. The advent of organoid formation presents an innovative avenue that may usher in a paradigm shift in diagnosing pancreatic cystic lesions.
AUTHOR CONTRIBUTIONS
Conceptualisation: Dongxi Xiang. Methodology: Fei Jiang, Dongyan Cao and Hui Jiang. Software: Dongyan Cao and Dongxi Xiang. Investigation: Zhendong Jin, Yingbin Liu and Dongxi Xiang. Formal analysis: Fei Jiang, Dongyan Cao and Gengming Niu. Writing—original draft: Fei Jiang, Dongyan Cao and Dongxi Xiang. Writing—review and editing: Dongxi Xiang. Visualisation: Fei Jiang and Dongyan Cao. Funding acquisition: Dongxi Xiang. Resources: Zhendong Jin, Yingbin Liu and Dongxi Xiang. Supervision: Dongxi Xiang.
ACKNOWLEDGEMENTS
The research was supported by grants from the Natural Science Foundation of China (32170924), the Shanghai Municipal Committee of Science and Technology (21140901600) and the Shanghai Jiaotong University School of Medicine (02.101005.001.29.38A). 2019 Climbing Peaks 234 Proect (2019YXK017).
CONFLICT OF INTEREST STATEMENT
Dr. Gengming Niu is an employee of Shanghai OneTar Biomedicine. The remaining authors declare they have no conflicts of interest.
ETHICS STATEMENT
All patients signed the informed consent forms and the study was approved by Renji Hospital Ethics Committee of Shanghai Jiaotong University School of Medicine (no. KY2023-034-B).
Open Research
DATA AVAILABILITY STATEMENT
To access future discoveries in this topic, we have uploaded the sequencing data to the NCBI and the matrices are available through GEO (GSE252896).




