Role of DNA methylation in cancer development and its clinical applications
Abstract
methyltransferase gene (DNA) methylation is a key process in epigenetic modification. This transformation leads to genomic instability that creates an impact on gene expression. DNA methylation is also involved in several mutations of tumour suppressor genes (TSGs) or proto-oncogenes, resulting in cancer (hypermethylation and hypomethylation). Hypermethylation of promoter segments in TSGs causes transcriptional silencing, whereas hypomethylation of regulatory sequence activates proto-oncogenes and retrotransposons. DNA methylation regulated by DNA methyltransferases is one of the essential epigenetic mechanisms that controls the cell cycle, cell proliferation, differentiation, apoptosis and transformation in eukaryotes, leading to genetic instability of tumour cells. Recent scientific research has highlighted that DNA methylation is a vital cancer biomarker source of several parts of body fluids that allow primary-stage cancer cell detection during clinical screening. Nowadays, epigenetic biomarkers have been introduced as a decision maker with the potential to improve cancer prognosis. DNA methylation profiling helps to determine cancer at the deep genetic level, and create high impactful cancer screening approach in the future.
1 INTRODUCTION
Cancer is the most challenging health crisis worldwide. Treatment of this disease is very difficult because early detection of cancer is more critical for clinical experts. The lack of advanced diagnostic tool development has an impact on the poor survival rate of cancer patients. In this circumstance, more promising diagnostic biomarkers are needed. Cancer originates from genetic instability of cells, which are dynamically regulated by epigenetic modification. DNA methylation based on several mutations leads to the development of carcinogenic health complications. This genetic reprogramming happens via inhibition or initiation of several signalling pathways.1, 2 Epigenetic alteration takes place early in the development of cancer, and because of that several research groups focus on more detailed research on this event for the invention of more promising cancer biomarkers for early detection.3 DNA methylation plays a vital role in gene expression, and alteration of DNA methylation patent promotes cancer progression.4 Based on this understanding, it may be a biomarker of cancer, and may also play a role in the development of promising therapeutic approaches.
2 DNA METHYLATION IN CANCER
DNA methylation is one of the vital cellular transcription regulatory events that also play a role in cancer development.5 Abnormal DNA methylation leads to the development of oncogenic properties in cells via hypomethylation or hypermethylation. This alteration is involved in epigenetic modifications. Hypermethylation is related to tumour suppressor gene silencing, and this transformation has identified the global reason behind oncogenes.6 In the case of cancer, DNA methylation mostly affects suppressed genes and activates oncogenes.
3 DNA METHYLTRANSFERASES
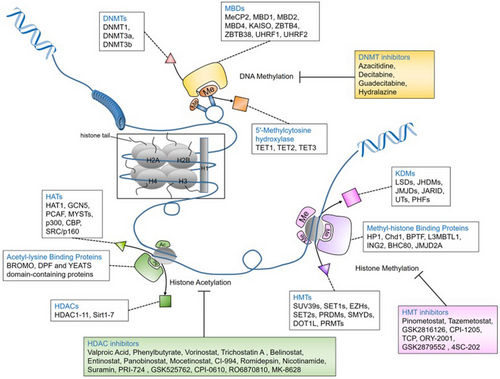
DNA methyltransferases (DNMTs) are a group of enzymes that are involved in DNA methylation.7, 8 These enzymes carry S-adenosyl methionine as a methyl group, which is transported into cytosine at the five positions of the pyrimidine ring. Humans contain four types of DNMTs (DNMT1, DNMT2, DNMT3a and DNMT3b). The role of DNMTs in DNA methylation is summarised in Table 1. DNA methylation and histone methylation-based epigenetic regulation are described in Figure 1.
| DNMTs | Function in DNA methylation | Involve in cancer |
|---|---|---|
| DNMT1 | It maintains methyl group in the semi-methylated stand of DNA during DNA replication |
Colon cancer, pancreas cancer, esophageal cancer |
| DNMT2 | It plays a role in cytosine methylation | Breast cancer |
| DNMT3a | It transports methyl group in specific CpG side of DNA | Acute myeloid leukaemia, lymphoma, liver cancer |
| DNMT3b | It is involved in de novo DNA methylation and works with H3K36me3 and RNA Pol-II | Acute myeloid leukaemia, prostate cancer, breast cancer |
4 ROLE OF DNA HYPOMETHYLATION IN CANCER DEVELOPMENT
Global scientific investigation has shown that hypomethylation is found in cancer cells and regulates the overexpression of proto-oncogenes and growth factors related to uncontrolled cellular growth, metastasis and genes.10 DNA hypomethylation is initiated by multiple factors such as tumour gene—PLAU, S100A4 and calcium-binding protein. All of this protein production modulated the function of the extracellular matrix as it developed the motility of the malignant cell population The aggressive phage of cancer cells produces protease that damages extracellular matrix and promotes cancer metastasis. The S100A4 protein regulated enzyme synthesis, which is related to healing of extracellular matrix damage.11 The PLAU enzyme plays a vital role in cancer metastasis of the prostate, breast and other parts of the body.10 Cancer cell proliferation is influenced by insulin-like growth factor 2 (IGF2). Inside tumour cells, hypomethylation alters the IGF2 gene allele; as a result, this event boosts the cancer cell division. The retrotransposons (long interspersed nuclear elements belong in this class)-based hypomethylation generate cancer cell's genetic instability. The precise reason behind DNA hypomethylation is still not clear.
5 ROLE OF DNA HYPERMETHYLATION IN CANCER
DNA hypermethylation occurs in several activated genes, as a result of this event, epigenetic silencing takes place and leads to cancer cell's genetic instability. The primary epigenetic modification invented the promoter part of the retinoblastoma tumour-suppressor gene (RB1). The hypermethylation leads to large population of tumour suppression gene silencing, which regulates tumourigenesis hallmark in human cancer. This activity in cells influences multiple-cell signalling. These two p15 and p16 cell cycle-related gene hypermethylations were observed in several cancers, both of which regulate the G1 phage of the cell cycle. Abnormal DNA methylation affects the DNA repair system in cells. For example, DNA mismatch repair mechanism-based hypermethylation is involved with gastric cancers and it also breaks DNA stand in the case of breast and ovarian cancer. Silencing of the CDH1 gene (E-cadherin) promotes the development of metastasis nature in cancer cells, and silencing of death-associated protein kinase 1 (DAPK1) gene leads to the development of cancer cell apoptosis escape property in cells. Current research highlighted one of the most interesting facts about DNA methylation in cancer—every cancer type and cell specifically carry a unique pattern.8
6 DNA METHYLATION PROMOTING CANCER DEVELOPMENT, INVASION AND METASTASES
DNA methylation-based chromosomal instability leads to the initiation of cancer metastasis. The epithelial–mesenchymal transition (EMT) is a major step in cancer metastases. In normal cases, epithelial cells are tightly bound with other cells via extracellular matrix (ECM) and during cancer development, ECM is modified; as a result, cells become motile and spread to different parts of the body. E-cadherin is the key factor in EMT; in cancer, it is downregulated. Multiple events take place to regulate the E-cadherin such as somatic mutation and DNA methylation. The chromosome 16-based DNA methylation plays a role in epigenetic modification of E-cadherin.12 Scientific investigation observed that in the case of breast cancer, mutation happens in 16q22.1 chromosomal location. The methylation of 5′ CpG (oral cancer, liver cancer, skin cancer, and blood cancer), CpG hypermethylation (bladder cancer) and p16 (lung cancer, kidney cancer) gene methylation is involved in aggressive cancer development.
7 DNA METHYLATION-BASED MARKER FOR CANCER
Current scenario of cancer research, based on understanding, highlighted that cancer requires innovative tools and effective diagnostic approaches. The biomarker development research ideology involves not only early detection of cancer but also therapeutic efficacy monitoring and post-surgery cancer risk investigation. DNA methylation-based biomarker development is more promising in cancer diagnosis.13 Moreover, this biomarker provides dynamic information about mutation at the cellular level, which involves several stages of cancer progression. The Breast cancer type 1 gene (BRCA1) (ovary cancer), secreted frizzled related protein 2 gene (SFRP2), MGMT (O-6-methylguanine-DNA methyltransferase) (colon), p16 (lung cancer) and p15 (blood cancer) methylation-based biomarkers were screened by liquid biopsy.
8 DNA METHYLATION PROFILING TECHNOLOGIES
A DNA methylation analysis-based epigenetic investigation was conducted globally for a more detailed cancer investigation. Generally, 5-methylcytosine (5mC) found in cancer may serve as a cancer biomarker. The vital discovery unfolds the misty of 5mC −5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine, which open up a dynamic chapter of epigenetic. Currently, several technologies14, 15 are being developed for DNA methylation platforms, which are explained in Table 2.
| Technologies | Function |
|---|---|
|
NGS methods apply for quantitative DNA methylation data at single base pair resolution and with genome-wide coverage |
|
|
|
Quantification of DNA methylation to be faster than other HPLC-based methods |
|
|
|
|
| High-throughput-based DNA methylation analysis |
|
|
The redox reactions-based DNA methylation |
|
|
|
|
|
|
|
|
- Abbreviations: HPCE, high-performance capillary electrophoresis; HPLC, high-performance liquid chromatography; NGS, next-generation sequencing; SERS, surface-enhanced Raman scattering; 5hmC, 5-hydroxymethylcytosine; 5mC, 5-methylcytosine
9 CLINICAL IMPORTANCE
The current advantage of cancer-associated DNA methylation research is the initiation of a new era for early cancer detection and improved drug efficacy.16 The DNA methylation base biomarker investigation is the most highlighted field now; in this purpose, multiple body fluids are analysed such as plasma, sputum, stool and urine. This process has several limitations in large-scale clinical applications. First, some DNA methylation happens at minute level; for this reason, more sensitive tool development is needed. Second, existing DNA methylation analysis assays are based on primary tumours, and this platform is not suitable for liquid biopsy. The revolutionary technical progress in the medical field may overcome this limitation with innovative approaches. Recent whole-genome methylation profiling technique provides dynamic information about thousands of loci, which helps us to identify more sensitive specific DNA methylation-based novel markers for cancer.
AUTHOR CONTRIBUTIONS
Swarup Sonar and Sidhanti Nyahatkar: Manuscript writing; Ketki Kalele: Reviewing; Manab Deb Adhikari: Final Reviewing and editing.
ACKNOWLEDGEMENTS
Not applicable.
CONFLICT OF INTEREST STATEMENT
The authors declare they have no conflicts of interest.
FUNDING INFORMATION
The authors received no specific funding for this work.
ETHICS STATEMENT
Not applicable




