Characterization of HpnG as a Purine Nucleoside Phosphorylase in Bacteriohopanepolyol Biosynthesis†
Xinhui Li
Department of Chemistry, Fudan University, Shanghai, 200433 China
These authors contributed equally to this work.
Search for more papers by this authorXiaoyu Zhu
Department of Chemistry, Fudan University, Shanghai, 200433 China
These authors contributed equally to this work.
Search for more papers by this authorYuting Zhong
Department of Chemistry, Fudan University, Shanghai, 200433 China
State Key Laboratory of Microbial Metabolism, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, Shanghai, 200240 China
Search for more papers by this authorWei Zhang
Key Laboratory of Extreme Environmental Microbial Resources and Engineering of Gansu Province, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, Gansu, 73000 China
Search for more papers by this authorFener Chen
Department of Chemistry, Fudan University, Shanghai, 200433 China
Search for more papers by this authorWenning Wang
Department of Chemistry, Fudan University, Shanghai, 200433 China
Search for more papers by this authorWei Ding
State Key Laboratory of Microbial Metabolism, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, Shanghai, 200240 China
Search for more papers by this authorCorresponding Author
Qi Zhang
Department of Chemistry, Fudan University, Shanghai, 200433 China
E-mail: [email protected]Search for more papers by this authorXinhui Li
Department of Chemistry, Fudan University, Shanghai, 200433 China
These authors contributed equally to this work.
Search for more papers by this authorXiaoyu Zhu
Department of Chemistry, Fudan University, Shanghai, 200433 China
These authors contributed equally to this work.
Search for more papers by this authorYuting Zhong
Department of Chemistry, Fudan University, Shanghai, 200433 China
State Key Laboratory of Microbial Metabolism, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, Shanghai, 200240 China
Search for more papers by this authorWei Zhang
Key Laboratory of Extreme Environmental Microbial Resources and Engineering of Gansu Province, Northwest Institute of Eco-Environment and Resources, Chinese Academy of Sciences, Lanzhou, Gansu, 73000 China
Search for more papers by this authorFener Chen
Department of Chemistry, Fudan University, Shanghai, 200433 China
Search for more papers by this authorWenning Wang
Department of Chemistry, Fudan University, Shanghai, 200433 China
Search for more papers by this authorWei Ding
State Key Laboratory of Microbial Metabolism, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, Shanghai, 200240 China
Search for more papers by this authorCorresponding Author
Qi Zhang
Department of Chemistry, Fudan University, Shanghai, 200433 China
E-mail: [email protected]Search for more papers by this authorDedicated to the Special Issue of Emerging Investigators in 2024.
Comprehensive Summary
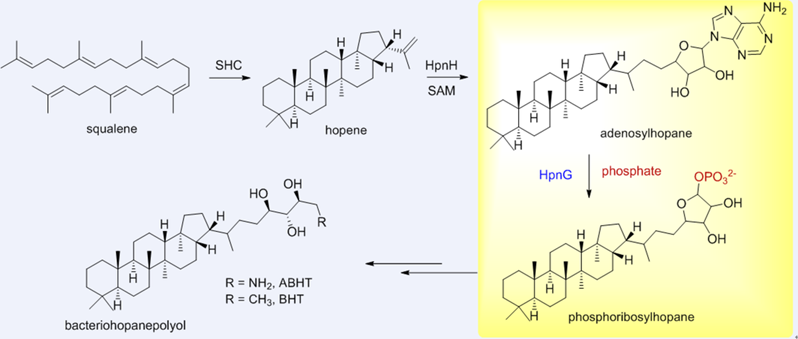
HpnG plays a crucial role in the production of ribosylhopane, a key intermediate in the biosynthesis of bacteriohopanepolyol. Despite early extensive studies, the precise function of HpnG has remained elusive. Here, we report functional characterization of HpnG as a purine nucleoside phosphorylase, which converts adenosylhopane to phosphoribosylhopane in the presence of phosphate. HpnG demonstrates broad substrate specificity and impressive stability, making it a valuable enzymatic tool for applications in nucleoside processing and related biotechnology.
Supporting Information
| Filename | Description |
|---|---|
| cjoc202400233-sup-0001-supinfo.pdfPDF document, 1.2 MB |
Appendix S1: Supporting Information |
Please note: The publisher is not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing content) should be directed to the corresponding author for the article.
References
- 1(a) Blumenberg, M.; Oppermann, B. I.; Guyoneaud, R.; Michaelis, W. Hopanoid production by Desulfovibrio bastinii isolated from oilfield formation water. FEMS Microbiol. Lett. 2009, 293, 73–78; (b) Cvejic, J. H.; Putra, S. R.; El-Beltagy, A.; Hattori, R.; Hattori, T.; Rohmer, M. Bacterial triterpenoids of the hopane series as biomarkers for the chemotaxonomy of Burkholderia, Pseudomonas and Ralstonia spp. FEMS Microbiol. Lett. 2000, 183, 295–299.
- 2(a) O'Beirne, M. D.; Sparkes, R.; Hamilton, T. L.; van Dongen, B. E.; Gilhooly Iii, W. P.; Werne, J. P. Characterization of diverse bacteriohopanepolyols in a permanently stratified, hyper-euxinic lake. Org. Geochem. 2022, 168, 104431; (b) Kusch, S.; Rush, D. Revisiting the precursors of the most abundant natural products on Earth: A look back at 30+ years of bacteriohopanepolyol (BHP) research and ahead to new frontiers. Org. Geochem. 2022, 172, 104469; (c) Garby, T. J.; Jordan, M.; Timms, V.; Walter, M. R.; Neilan, B. A. 2-Methylhopanoids in geographically distinct, arid biological soil crusts are primarily cyanobacterial in origin. Environ. Microbiol. Rep. 2022, 14, 164–169; (d) Elling, F. J.; Evans, T. W.; Nathan, V.; Hemingway, J. D.; Kharbush, J. J.; Bayer, B.; Spieck, E.; Husain, F.; Summons, R. E.; Pearson, A. Marine and terrestrial nitrifying bacteria are sources of diverse bacteriohopanepolyols. Geobiology 2022, 20, 399–420; (e) Matys, E. D.; Mackey, T.; Grettenberger, C.; Mueller, E.; Sumner, D. Y.; Hawes, I.; Summons, R. E. Bacteriohopanepolyols across environmental gradients in Lake Vanda, Antarctica. Geobiology 2019, 17, 308–319; (f) Matys, E. D.; Mackey, T.; Grettenberger, C.; Mueller, E.; Jungblut, A.; Sumner, D. Y.; Hawes, I.; Summons, R. E. Environmental controls on bacteriohopanepolyol profiles of benthic microbial mats from Lake Fryxell, Antarctica. Geobiology 2019, 17, 551–563.
- 3 Kusch, S.; Walter, S. R. S.; Hemingway, J. D.; Pearson, A. Improved chromatography reveals multiple new bacteriohopanepolyol isomers in marine sediments. Org. Geochem. 2018, 124, 12–21.
- 4(a) Gonzalez-Rosales, C.; Vergara, E.; Dopson, M.; Valdes, J. H.; Holmes, D. S. Integrative Genomics Sheds Light on Evolutionary Forces Shaping the Acidithiobacillia Class Acidophilic Lifestyle. Front. Microbiol. 2022, 12, 822229; (b) Rizk, S.; Henke, P.; Santana-Molina, C.; Martens, G.; Gnaedig, M.; Nguyen, N. A.; Devos, D. P.; Neumann-Schaal, M.; Saenz, J. P. Functional diversity of isoprenoid lipids in Methylobacterium extorquens PA1. Mol. Microbiol. 2021, 116, 1064–1078; (c) Kolouchova, I.; Timkina, E.; Matatkova, O.; Kyselova, L.; Rezanka, T. Analysis of Bacteriohopanoids from Thermophilic Bacteria by Liquid Chromatography-Mass Spectrometry. Microorganisms 2021, 9, 2062; (d) S. Alvares, D.; Crosio, M.; Wilke, N. Hopanoid Hopene Locates in the Interior of Membranes and Affects Their Properties. Langmuir 2021, 37, 11900–11908; (e) Chwastek, G.; Surma, M. A.; Rizk, S.; Grosser, D.; Lavrynenko, O.; Rucinska, M.; Jambor, H.; Saenz, J. Principles of Membrane Adaptation Revealed through Environmentally Induced Bacterial Lipidome Remodeling. Cell Rep. 2020, 32, 108165; (f) Rivas-Marin, E.; Stettner, S.; Gottshall, E. Y.; Santana-Molina, C.; Helling, M.; Basile, F.; Ward, N. L.; Devos, D. P. Essentiality of sterol synthesis genes in the planctomycete bacterium Gemmata obscuriglobus. Nat. Commun. 2019, 10, 2916; (g) Lodha, T. D.; Indu, B.; Sasikala, C.; Ramana, C. V. Transcriptome analysis of hopanoid deficient mutant of Rhodopseuodomonas palustris TIE-1. Microbiol. Res. 2019, 218, 108–117; (h) Brenac, L.; Baidoo, E. E. K.; Keasling, J. D.; Budin, I. Distinct functional roles for hopanoid composition in the chemical tolerance of Zymomonas mobilis. Mol. Microbiol. 2019, 112, 1564–1575; (i) Belin, B. J.; Tookmanian, E. M.; de Anda, J.; Wong, G. C. L.; Newman, D. K. Extended Hopanoid Loss Reduces Bacterial Motility and Surface Attachment and Leads to Heterogeneity in Root Nodule Growth Kinetics in a Bradyrhizobium-Aeschynomene Symbiosis. Mol. Plant. Microb. Interact. 2019, 32, 1415–1428.
- 5(a) Welander, P. V.; Doughty, D. M.; Wu, C. H.; Mehay, S.; Summons, R. E.; Newman, D. K. Identification and characterization of Rhodopseudomonas palustris TIE-1 hopanoid biosynthesis mutants. Geobiology 2012, 10, 163–177; (b) Bradley, A. S.; Pearson, A.; Saenz, J. P.; Marx, C. J. Adenosylhopane: The first intermediate in hopanoid side chain biosynthesis. Org. Geochem. 2010, 41, 1075–1081; (c) Liu, W.; Sakr, E.; Schaeffer, P.; Talbot, H. M.; Donisi, J.; Hartner, T.; Kannenberg, E.; Takano, E.; Rohmer, M. Ribosylhopane, a novel bacterial hopanoid, as precursor of C35 bacteriohopanepolyols in Streptomyces coelicolor A3(2). ChemBioChem 2014, 15, 2156–2161; (d) Challis, G. L. Exploitation of the Streptomyces coelicolor A3(2) genome sequence for discovery of new natural products and biosynthetic pathways. J. Ind. Microbiol. Biotechnol. 2014, 41, 219–232.
- 6(a) Bouwknegt, J.; Wiersma, S. J.; Ortiz-Merino, R. A.; Doornenbal, E. S. R.; Buitenhuis, P.; Giera, M.; Mueller, C.; Pronk, J. T. A squalene-hopene cyclase in Schizosaccharomyces japonicus represents a eukaryotic adaptation to sterol-limited anaerobic environments. Proc. Natl. Acad. Sci. 2021, 118, e2105225118; (b) Santana-Molina, C.; Rivas-Marin, E.; M. Rojas, A.; Devos, D. P. Origin and Evolution of Polycyclic Triterpene Synthesis. Mol. Biol. Evol. 2020, 37, 1925–1941; (c) Pattanaik, B.; Englund, E.; Nolte, N.; Lindberg, P. Introduction of a green algal squalene synthase enhances squalene accumulation in a strain of Synechocystis sp. PCC 6803. Metab. Eng. Commun. 2020, 10, e00125; (d) Nair, I. M.; Jayachandran, K. 4–4' Diaponeurosporenic Acid, the C-30 Carotenoid Pigment in Endophytic Pseudomonas Mendocina with Squalene Cyclase Activity. Curr. Microbiol. 2020, 77, 3473–3479; (e) Liu, Z.; Zhang, Y.; Sun, J.; Huang, W.-C.; Xue, C.; Mao, X. A Novel Soluble Squalene-Hopene Cyclase and Its Application in Efficient Synthesis of Hopene. Front. Bioeng. Biotechnol. 2020, 8, 426; (f) Nakano, C.; Watanabe, T.; Minamino, M.; Hoshino, T. Enzymatic syntheses of novel carbocyclic scaffolds with a 6,5+5,5 ring system by squalene-hopene cyclase. Org. Biomol. Chem. 2019, 17, 9375–9389; (g) Ma, K.; Zhang, P.; Tao, Q.; Keller, N. P.; Yang, Y.; Yin, W.-B.; Liu, H. Characterization and Biosynthesis of a Rare Fungal Hopane-Type Triterpenoid Glycoside Involved in the Antistress Property of Aspergillus fumigatus. Org. Lett. 2019, 21, 3252–3256.
- 7(a) Sato, S.; Kudo, F.; Rohmer, M.; Eguchi, T. Biochemical and Mutational Analysis of Radical S-Adenosyl-L-Methionine Adenosylhopane Synthase HpnH from Zymomonas mobilis Reveals that the Conserved Residue Cysteine-106 Reduces a Radical Intermediate and Determines the Stereochemistry. Biochemistry 2021, 60, 2865–2874; (b) Hopmans, E. C.; Smit, N. T.; Schwartz-Narbonne, R.; Damste, J. S. S.; Rush, D. Analysis of non-derivatized bacteriohopanepolyols using UHPLC-HRMS reveals great structural diversity in environmental lipid assemblages. Org. Geochem. 2021, 160, 104285; (c) Zhong, Y.; Ji, X.; Zhang, Q. Radical SAM-Dependent Adenosylation Involved in Bacteriohopanepolyol Biosynthesis. Chin. J. Chem. 2020, 38, 39–42; (d) Sato, S.; Kudo, F.; Rohmer, M.; Eguchi, T. Characterization of Radical SAM Adenosylhopane Synthase, HpnH, which Catalyzes the 5'-Deoxyadenosyl Radical Addition to Diploptene in the Biosynthesis of C-35 Bacteriohopanepolyols. Angew. Chem. Int. Ed. 2020, 59, 237–241.
- 8 Blin, K.; Shaw, S.; Augustijn, H. E.; Reitz, Z. L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B. R.; Metcalf, W. W.; Helfrich, E. J. N.; van Wezel, G. P.; Medema, M. H.; Weber, T. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structuresand visualisation. Nucleic Acids Res. 2023, 51, W46–W50.
- 9 Bzowskaa, A.; Kulikowskaa, E.; Shugar, D. Purine nucleoside phosphorylases: properties, functions, and clinical aspects. Pharmacol. Therapeut. 2000, 88, 349–425.
- 10 Ngivprom, U.; Kluaiphanngam, S.; Ji, W. J.; Siriwibool, S.; Kamkaew, A.; Cairns, J. R. K.; Zhang, Q.; Lai, R. Y. Characterization of NucPNP and NucV involved in the early steps of nucleocidin biosynthesis in Streptomyces calvus. RSC Adv. 2021, 11, 3510–3515.
- 11 Xu, G.; Kong, L.; Gong, R.; Xu, L.; Gao, Y.; Jiang, M.; Cai, Y. S.; Hong, K.; Hu, Y.; Liu, P.; Deng, Z.; Price, N. P. J.; Chen, W. Coordinated Biosynthesis of the Purine Nucleoside Antibiotics Aristeromycin and Coformycin in Actinomycetes. Appl. Environ. Microbiol. 2018, 84, e01860–01818.
- 12 Joshi, S.; Fedoseyenko, D.; Mahanta, N.; Manion, H.; Naseem, S.; Dairi, T.; Begley, T. P. Novel enzymology in futalosine-dependent menaquinone biosynthesis. Curr. Opin. Chem. Biol. 2018, 47, 134–141.
- 13(a) Narczyk, M.; Wojtys, M. I.; Asler, I. L.; Zinic, B.; Luic, M.; Jagusztyn-Krynicka, E. K.; Stefanic, Z.; Bzowska, A. Interactions of 2,6-substituted purines with purine nucleoside phosphorylase from Helicobacter pylori in solution and in the crystal, and the effects of these compounds on cell cultures of this bacterium. J. Enzyme Inhibit. Med. Chem. 2022, 37, 1083–1097; (b) Dziekan, J. M.; Yu, H.; Chen, D.; Dai, L.; Wirjanata, G.; Larsson, A.; Prabhu, N.; Sobota, R. M.; Bozdech, Z.; Nordlund, P. Identifying purine nucleoside phosphorylase as the target of quinine using cellular thermal shift assay. Sci. Transl. Med. 2019, 11, eaau3174; (c) Furman, R. R.; Hoelzer, D. Purine nucleoside phosphorylase inhibition as a novel therapeutic approach for B-cell lymphoid malignancies. Semin. Oncol. 2007, 34, S29–S34; (d) Pereira, H. D.; Franco, G. R.; Cleasby, A.; Garratt, R. C. Structures for the potential drug target purine nucleoside phosphorylase from Schistosoma mansoni causal agent of schistosomiasis. J. Mol. Biol. 2005, 353, 584–599.
- 14 Cantalapiedra, C. P.; Hernandez-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829.
- 15 Tozzi, M. G.; Camici, M.; Mascia, L.; Sgarrella, F.; Ipata, P. L. Pentose phosphates in nucleoside interconversion and catabolism. FEBS J. 2006, 273, 1089–1101.
- 16(a) Wu, R.; Ding, W.; Zhang, Q. Consecutive Methylation catalyzed by TsrM, an atypical Class B radical SAM methylase. Chin. J. Chem. 2022, 40, 1693–1698; (b) Cheng, J.; Ding, W.; Zhang, Q. Radical SAM-dependent Demetallation of Heme. Chin. J. Chem. 2022, 40, 1053–1058; (c) Bennett, M. R.; Shepherd, S. A.; Cronin, V. A.; Micklefield, J. Recent advances in methyltransferase biocatalysis. Curr. Opin. Chem. Biol. 2017, 37, 97–106; (d) Broderick, J. B.; Duffus, B. R.; Duschene, K. S.; Shepard, E. M. Radical S-adenosylmethionine enzymes. Chem. Rev. 2014, 114, 4229–4317.
- 17 Parveen, N.; Cornell, K. A. Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol. Microbiol. 2011, 79, 7–20.
- 18 Gericke, L.; Mhaindarkar, D.; Karst, L. C.; Jahn, S.; Kuge, M.; Mohr, M. K. F.; Gagsteiger, J.; Cornelissen, N. V.; Wen, X.; Mordhorst, S.; Jessen, H. J.; Rentmeister, A.; Seebeck, F. P.; Layer, G.; Loenarz, C.; Andexer, J. N. Biomimetic S-Adenosylmethionine Regeneration Starting from Multiple Byproducts Enables Biocatalytic Alkylation with Radical SAM Enzymes. ChemBioChem 2023, 24, e202300133.
- 19(a) Savarese, T. M.; Crabtree, G. W.; Parks, R. E. 5'-Methylthioadenosine phosphorylase .1. substrate activity of 5'-deoxyadenosine with the enzyme from sarcoma 180 cells. Biochem. Pharmacol. 1981, 30, 189–199; (b) North, J. A.; Wildenthal, J. A.; Erb, T. J.; Evans, B. S.; Byerly, K. M.; Gerlt, J. A.; Tabita, F. R. A bifunctional salvage pathway for two distinct S-adenosylmethionine by-products that is widespread in bacteria, including pathogenic Escherichia coli. Mol. Microbiol. 2020, 113, 923–927.
- 20(a) Tu, Y.; Li, H.; Tu, T.; Zhang, Q. Lamellar Enzyme-Metal–Organic Framework Composites Enable Catalysis on Large Substrates. CCS Chem. 2021, 4, 872–879;
10.31635/ccschem.021.202000759 Google Scholar(b) Li, H.; Tu, Y.; Tu, T.; Zhang, Q. Immobilization of Proteases on Nanoflower-Like Metal Organic Framework. Chin. J. Chem. 2023, 41, 1504–1508..




