Two distinct stem cell-like subtypes of hepatocellular carcinoma with clinical significance and their therapeutic potentials
Abbreviations
-
- BCCP
-
- Bayesian compound covariate prediction
-
- BET
-
- bromodomain and extra-terminal domain
-
- dHCC
-
- differentiated HCC
-
- HCC
-
- hepatocellular carcinoma
-
- HS
-
- hepatic stem cells
-
- HS1
-
- hepatic stem cell 1
-
- HS2
-
- hepatic stem cell 2
-
- PDX
-
- patient-derived xenograft
-
- TIDE
-
- tumor immune dysfunction and exclusion
Hepatocellular carcinoma (HCC) is among the most common cancers worldwide, causing about 600,000 deaths annually [1]. In HCC, stem cell-like characteristics, which drive early recurrence and therapy resistance, are major contributors to poor prognosis [2]. In this current study, we integrated and analyzed gene expression data from human fetal liver cells and primary HCC tumors (n = 1231) and uncovered two clinically and biologically distinct hepatic stem cell (HS) subtypes, potential biomarkers associated with these subtypes, and a potential new therapeutic intervention for these subtypes.
By analyzing gene expression data from human fetal liver cells [3], we identified 609-, 2538-, and 1139-gene signatures for gestational 10-week fetal liver cells, 17-week fetal liver cells, and mature hepatocytes, respectively (Supplementary Fig. S1A). Because 10- and 17-week fetal liver cells reflect different degrees of stemness of hepatic lineage, as reflected by α-fetoprotein expression (Supplementary Fig. S1B), we renamed the gene signatures specific to the 10- and 17-week cells hepatic stem cell type 1 (HS1) and hepatic stem cell type 2 (HS2), respectively. To estimate the clinical relevance of stem cell-like characteristics in primary HCC, we trichotomized HCC tumors as HS1, HS2, or differentiated HCC (dHCC) subtypes according to their degree of stemness by applying the prediction algorithm to the gene expression data from HCC tumors (n = 1226) (Supplementary Fig. S1C). The HS1 subtype was associated with worst overall survival, the HS2 subtype exhibited moderate overall survival, and the dHCC subtype exhibited the best overall survival (P < 0.001, Figure 1A). When patients with Barcelona Clinic Liver Cancer stage A or B disease were stratified by HS subtype, the subtypes successfully stratified high-risk patients at both stages (Supplementary Fig. S2). In addition to clinical stage and resection status, the HS subtypes were significant predictors of overall survival, except HS2 for recurrence-free survival, in multivariate Cox proportional hazards regression analyses (Supplementary Table S1 and S2). The HS subtypes were also significantly associated with some clinicopathological variables (Supplementary Fig. S3).
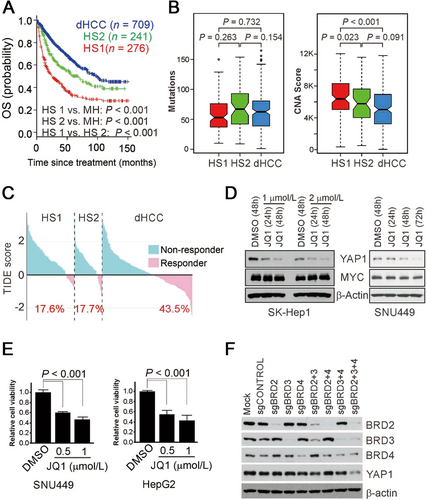
Clinical and biological significance of hepatic stem cell subtypes. (A) Kaplan-Meier plot of the OS for patients with hepatocellular carcinoma (HCC; n = 1226). HCC patients were trichotomized by Bayesian compound covariate prediction. P-values were estimated using log-rank tests. (B) Non-synonymous mutation rates and copy number alterations in stem cell subtypes of HCC in The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA-LIHC) cohort (n = 371). (C) Waterfall plots showing the rates of immunotherapy response predicted by the tumor immune dysfunction and exclusion (TIDE) algorithm in the TCGA-LIHC cohort. The percentages of patients with each subtype who were responders are shown below the plots. (D) Western blots of YAP1 and MYC after JQ1 treatment. The bromodomain and extra-terminal domain (BET) inhibitor JQ1 downregulated YAP1 expression but not MYC expression in SK-Hep1 and SNU449 cells. (E) Viability of HCC cells treated with JQ1. P-values were estimated using the Student t-test. (F) The depletion of three BET proteins (BRD2, BRD3, and BRD4) was necessary for the suppression of YAP1 expression in HepG2 cells. BET protein expression was depleted by single-guide RNAs specific to each BET protein.
Abbreviations: OS, overall survival; HCC, hepatocellular carcinoma; dHCC, differentiated hepatocytes; HS2, hepatic stem cell type 2; HS1, hepatic stem cell type 1; TCGA-LIHC, The Cancer Genome Atlas Liver Hepatocellular Carcinoma; TIDE, tumor immune dysfunction and exclusion; BET, bromodomain and extraterminal domain; sgBRD, single-guide RNA against BRD mRNAs; DMSO, dimethyl sulfoxide
Interestingly, while there was no significant difference in the subtypes’ mutation rates, HS1 subtype showed significantly different copy number alteration score when compared with other subtypes (Figure 1B). The HS1 subtype showed higher rates of TP53 and RB1 mutations, while the HS2 subtype showed frequent IL6ST and CDKN2A mutations (Supplementary Fig. S4). Since the HS1 subtype showed the highest microRNA expression (Supplementary Fig. S5), it will be interesting to test in future studies if they play important roles in maintaining the stemness of cancer cells.
As a combination of targeted treatment and immune checkpoint blockade has been reported to have encouraging results in HCC [4], we assessed each subtype's potential response to immunotherapy using tumor immune dysfunction and exclusion (TIDE) scores which reflects their degrees for immunotherapy resistance [5]. Both HS subtypes showed high TIDE scores (Figure 1C), suggesting that patients with either HS subtype would not benefit substantially from immunotherapy. In agreement with this, the HS subtype probability and TIDE scores were positively correlated (Supplementary Fig. S6A). In an analysis using the CIBERSORT algorithm [6], which estimates the relative fractions of immune cells (Supplementary Fig. S6B), the non-activated naïve M0 macrophage fraction was significantly higher in both HS subtypes (Supplementary Fig. S6C, D), suggesting that a lack of active macrophages could be contributing to the HS subtypes’ low immunotherapy response. Furthermore, the fraction of immunosuppressive regulatory T cells was higher in the HS subtypes (Supplementary Fig. S6C, D), suggesting that regulatory T cells also play roles in the immune reactivity of HCC cells. The level of myeloid-derived suppressor cells as determined by the TIDE algorithm was significantly elevated in both HS subtypes (Supplementary Fig. S7A). Likewise, the expression of major immune checkpoints (PD-1, PD-L1, and CTLA4) was significantly different in the HS subtypes (Supplementary Fig. S7B, C), further supporting the notion that the HS subtypes are immune-suppressive. Interestingly, the HS1 subtype was associated with potential response to sorafenib, while the HS2 subtype was not (Supplementary Fig. S8) (see extended discussion in Supplementary File).
We next applied the HS signatures to the gene expression data of 75 patient-derived xenograft (PDX) HCC tumors. The HS signatures of the PDX tumors are highly similar to primary HCC (Supplementary Fig. S9A), indicating that stem cell features are well-maintained in PDX tumors. Most PDX tumors appeared to be stable as the passage number of the PDX models differed only slightly among subtypes (Supplementary Fig. S9B). Using targeted ELISA for 122 serum proteins (Oncology MAP screening), we performed a proteomic analysis with serum samples from HCC patients (n = 45, a subset of our cohort) and identified the top five serum markers for the HS1 and HS2 subtypes (Supplementary Fig. S10).
Gene network analysis revealed that oncogenes were activated in the HS1 and HS2 subtypes (Supplementary Table S3 and S4). In particular, YAP1 was highly activated in the HS1 subtype. Since YAP1 regulates HS and is associated with poor prognosis in HCC [7], we next examined the potential interaction of other transcription regulators with YAP1 by integrating the downstream target genes of each of the transcription regulators. BRD4 exhibited the highest interaction with YAP1 as it shared more target genes with YAP1 than any of the other regulators (Supplementary Fig. S11A). Of BRD4’s 37 target genes, 15 (41%) were shared with YAP1 (Supplementary Fig. S11B), suggesting that BRD4 interacts with or regulates YAP1 in HCC. We also evaluated the expression of HCC-associated stem cell markers and found that many of these stem cell markers, including SALL4, were highly expressed in the HS1 subtype (Supplementary Fig. S12).
BRD4 is a member of the bromodomain and extraterminal domain (BET) family and a key transcriptional regulator for many oncogenes, including MYC [8]. Next, we tested if BET members were involved in the regulation of YAP1 expression. HCC cells treated with the BET family inhibitor JQ1 had reduced expression of YAP1 but not MYC, a de facto target of BET proteins (Figure 1D, Supplementary Fig. S13A) [8], suggesting that the oncogenic activity of the BET family is mediated by the regulation of YAP1 in HCC cells. Furthermore, JQ1 significantly reduced the viability and migration of HCC cells (Figure 1E, Supplementary Fig. S13B, C), suggesting that JQ1 can inhibit the growth and invasion of HCC cells by suppressing YAP1.
Since JQ1 could inhibit most BET family members, we silenced the expression of BET members to identify the key members regulating YAP1. Surprisingly, complete YAP1 suppression required the depletion of all BET members (Figure 1F), suggesting that all BET members are active and functional in HCC cells. Most HCC cells consistently expressed all BET members (Supplementary Fig. S13D). In agreement with this, BET family member gene expression was highly correlated with YAP1 expression in human HCC tumors (Supplementary Fig. S13E), which supports the notion that BET genes regulate YAP1 in HCC.
Previous studies identified HCC subtypes with stem cell features [9, 10] but provided only general descriptions of those features while also listing potential therapeutic targets without providing functional validation or guidance for treatment. In the current study, we found that HCCs with stem cell features were not clinically or genomically homogeneous. They differed not only in their degree of stemness but also in their clinical outcomes and underlying biology. Importantly, the HS1 subtype, which has a poor prognosis, appears to be sensitive to BET inhibitors. The newly identified serum markers associated with these subtypes may provide opportunities to develop marker-based clinical trials. Furthermore, the potential marker genes we identified are well-preserved in PDX models, which shows promise for the development of accurate disease models for preclinical study.
DECLARATIONS
AUTHORS' CONTRIBUTIONS
S.H.L., Y.S.J., and J.S.L. conceived the study, performed the literature search and bioinformatics analysis, and prepared the figures. S.L., B.H.S., H.K.H., G.H.C., C.M.K., J.S.C., W.J.L., J.H.C., A.K., L.W., S.Y.Y., Y.S.C., and H.J.J., helped with data collection, analysis, and interpretation. S.H.L., Y.S.J., and J.L. wrote and revised the manuscript. S.H.L and Y.S.J. contributed equally to this work. All authors read and approved the final manuscript.
ETHICAL APPROVAL AND CONSENT TO PARTICIPATE
Serum samples were obtained from HCC patients who provided informed consent under an IRB approved protocol at Mayo Clinic, Rochester, Minnesota, USA.
CONSENT FOR PUBLICATION
Not applicable
AVAILABILITY OF DATA AND MATERIALS
TCGA Data Portal: https://portal.gdc.cancer.gov/; Santa Cruz, Genomics Institute: https://xenabrowser.net; GEO Datasets for gene expression data sets GSE96981, GSE1898, GSE14520, GSE54236, GSE36376, GSE16757, and GSE43619: https://www.ncbi.nlm.nih.gov/gds/; All other data are available in supplemental information and original articles published earlier as described in the supplementary information.
FUNDING
This study was supported in part by National Cancer Institute grants R01-CA237327 and P50-CA217674, the Duncan Cancer Prevention Research Seed Funding Program at MD Anderson Cancer Center (2016 cycle), the MD Anderson Sister Institution Network Fund (2016 and 2019 cycles), and the National Institutes of Health through MD Anderson's Cancer Center Support Grant, P30-CA016672. Acquisition of blood samples was supported by grants R01-CA165076, P30-CA015083 (Survey Research Shared Resource at Mayo Clinic Cancer Center), and P50-CA210964 (Mayo Clinic Hepatobiliary SPORE, to L.R.R.). S.H.L. was supported by the Severance Research Initiative (SRI) project in Yonsei University College of Medicine.
COMPETING INTERESTS
The authors report no conflicts of interest.
ACKNOWLEDGMENT
We thank Joseph A Munch, Stephanie Deming, and Ashli Nguyen-Villarreal in MD Anderson's Research Medical Library for editing the manuscript.




